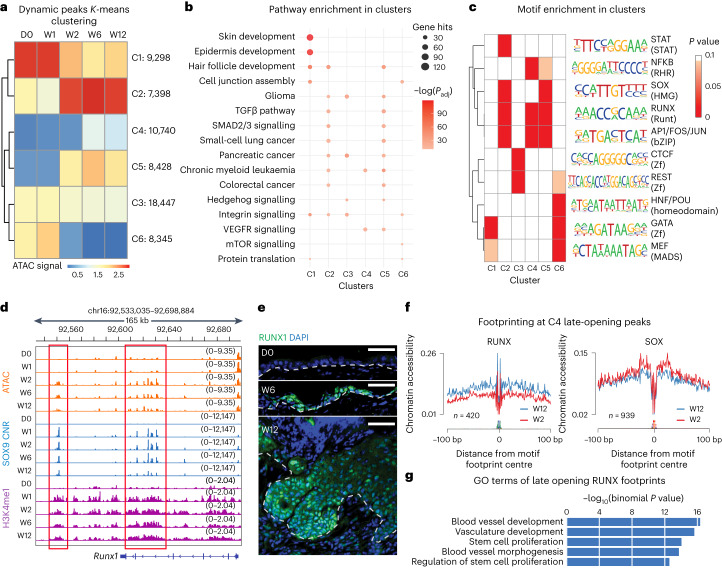
Fig. 3. SOX9 triggers an activated HFSC fate, while its target gene transcription factors trigger BCC transformation.
a, Heat map of K-means clustering of ATAC peaks based on signal across time. Number of peaks in each cluster are indicated on the right. b, GO-term analysis of the genes associated with each peak of a cluster. Size of circle reflects the number of gene hits for each pathway, while the shade of red indicates adjusted P value (Padj) from binomial test. Note that cluster C1, whose chromatin is suppressed by W2, is enriched for epidermal genes, while C2, whose chromatin is accessible by W2, is enriched for hair follicle genes. C4, whose chromatin is accessible at later times, is the cluster most enriched for BCC genes. c, Motif enrichment analysis of each cluster with HOMER. Shade of red indicates P value with white representing ≥0.01 (binomial is used to calculate the significance). Note enrichment for: GATA motif in epidermal genes whose chromatin closes after SOX9 induction; SOX motif in hair follicle genes whose chromatin opens early; and the SOX9 target RUNX1 motif in genes whose chromatin opens weeks after SOX9 has bound. d, Chromatin landscape of the Runx1 gene locus, showing that pioneer factor SOX9 binds to this gene (blue), concomitant with early increases in H3K4me1 modifications across the locus (purple), while chromatin accessibility (orange) comes afterwards. Red boxes indicate regions that are bound by SOX9, primed at W1 and opened at W2. e, RUNX1 immunofluorescence reveals its absence in epidermal homeostasis, but its presence at late timepoints (W6 and W12) following SOX9 induction. Scale bars, 50 μm. f, RUNX and SOX ATAC footprint analyses at W12 and W2, showing a late increase in chromatin accessibility at the RUNX footprint at a stage when phenotypic BCC-like invaginations are prevalent. By comparison, SOX footprints appear by W2 and are retained thereafter. g, GO terms of genes whose putative enhancers have RUNX footprints and open at W6 and W12 (binomial is used to calculate the significance).