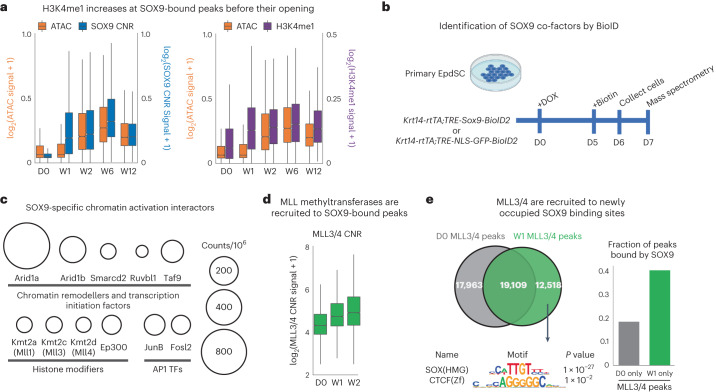
Fig. 4. SOX9 recruits co-factors to epigenetically prime and then remodel chromatin to an open, transcriptionally accessible state.
a, Left: box plot comparing ATAC (orange) and SOX9 CNR (blue) signals at SOX9 peaks that transition from closed to open chromatin over time. Note that SOX9 binding increases markedly by W1, preceding chromatin accessibility at these sites by nearly a week. Right: box plot comparing ATAC (orange) and MINT-ChIP H3K4me1 (purple) signals at SOX9 peaks that open over time. Note that H3K4me1 follows the time course of SOX9 binding, again preceding chromatin accessibility. n = 2 biological replicates. Box plots are centred at median and bound by first and the third quartile, and whiskers extend to 1.5 times interquartile range (IQR) on both ends. b, Schematic of BioID2 proximity labelling of proteins that interact with SOX9 induced in cultured EpdSCs. c, Selected SOX9-interacting proteins detected with mass spectrometry and that fall into the top GO terms of chromatin remodellers of the SWI/SNF family; transcription initiation factors (TAF9), AP1 transcription factors; and enzymes that modify histones (MLL1, MLL3, MLL4 and p300). For full list, see Supplementary Table 3. Circle size corresponds to the strength of the hit as delineated at right. d, Box plot of MLL3/4 CNR signals reveals the appearance of MLL3/4 at SOX9-bound peaks following DOX. n = 2 biological replicates. Box plot is centred at median and bound by the first and the third quartile, and whiskers extend to 1.5 times IQR on both ends. e, Venn diagrams providing further evidence that the de novo MLL3/4 peaks that appear between D0 and W1 are highly enriched for bound SOX9 (binomial P = 1 × 10−27), whereas the 17963 MLL3/4 peaks lost at W1 do not show any significant motif enrichment over all D0 MLL3/4 peaks and are not associated with SOX9-bound sites, inconsistent with a repressor role for SOX9.