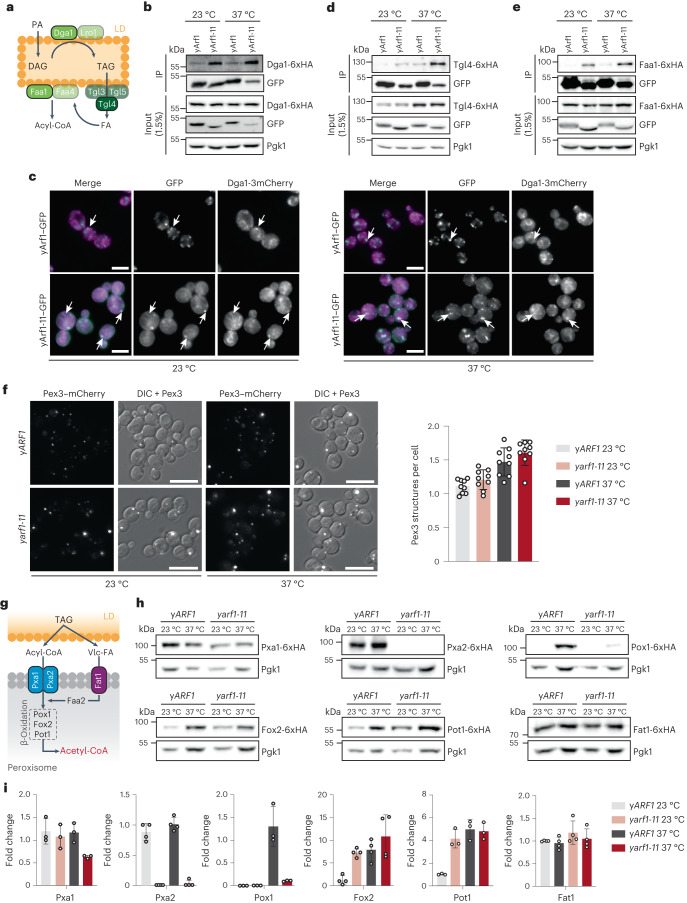
Fig. 7. yArf1 regulates LD-associated functions and β-oxidation.
a, Schematic of TAG synthesis and breakdown on LD. Key enzymes involved in TAG synthesis (Dga1), hydrolysis (Tgl4), and FA activation (Faa1) used in our co-IP experiments are shown in green. b,d,e, Co-IP of yArf1–GFP and yArf1-11–GFP with Dga1-6xHA (b), Tgl4-6xHA (d) and Faa1-6xHA (e). Strains were grown at 23 °C or shifted to 37 °C. c, Co-localization of yArf1–GFP and yArf1-11–GFP with the diacylglycerol acyltransferase Dga1 tagged with 3xmCherry grown at 23 °C or shifted to 37 °C for 30 min. Arrows indicate sites of co-localization or juxtaposition between the yArf1/yArf1-11 and Dga1 on the ER or on LD. Scale bars, 5 µm. f, Peroxisome biogenesis followed by microscopy using the peroxisomal marker Pex3 fused to mCherry in the yARF1 and yarf1-11 strains grown at 23 °C or shifted to 37 °C (left). Quantification of peroxisomes per cell in each strain and condition (right). Mean and standard deviation are shown; yARF1 + Pex3–mCherry 23 °C = 1,496 cells, yARF1 + Pex3–mCherry 37 °C = 1,070 cells, yarf1-11 + Pex3–mCherry 23 °C = 1,025 cells, yarf1-11 + Pex3–mCherry 23 °C = 970 cells from n = 3 biological replicates. Scale bars, 5 µm. g, Schematic of TAG mobilization to synthesize acetyl-CoA by peroxisomal β-oxidation in yeast. Relevant proteins monitored in h are shown. Vlc-FA, very-long-chain FAs. h, Immunoblot analysis of all β-oxidation proteins, both acyl-CoA transporters and the Vlc-FA transporter genomically fused to 6xHA in the yARF1 and yarf1-11 strains grown at 23 °C or shifted to 37 °C. Pgk1 was used as loading control. i, Relative fold changes in protein levels from immunodetections done in h. Mean and standard deviation are shown from n = 3 biological replicates. Source numerical data and unprocessed blots are available in source data. See also Extended Data Fig. 5.