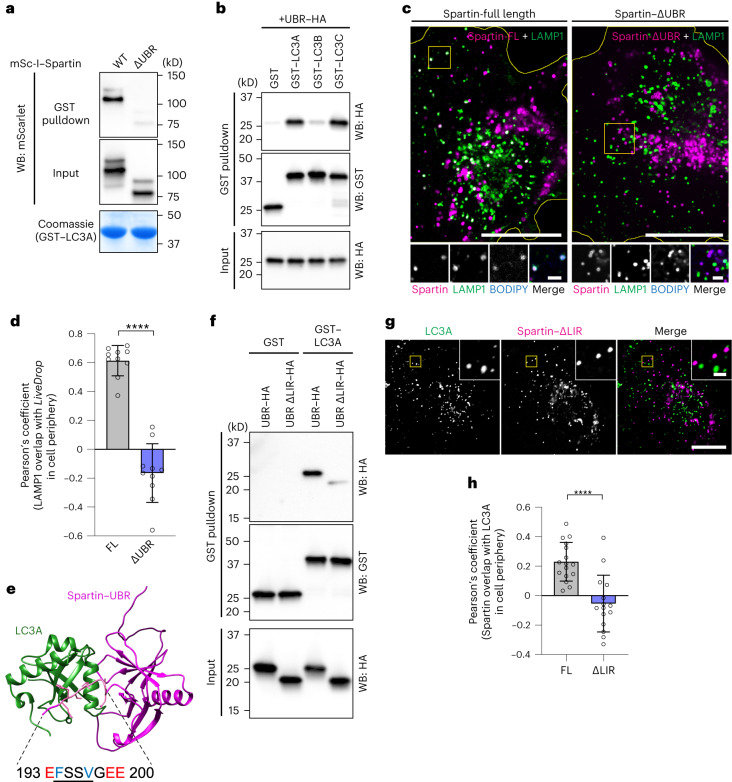
Fig. 3. UBR domain contains an LIR motif responsible for the interaction between spartin and LC3.
a, UBR domain is required for the interaction between spartin and LC3A. HEK293T WT cell lysates transiently expressing mScarlet-I–spartin WT or ΔUBR were incubated with recombinant GST–LC3A, followed by GST pulldown and detected with anti-mScarlet/mCherry antibody. Cells were treated will 0.5 mM OA for 24 h before lysate preparation. b, Recombinant UBR domain directly interacts with recombinant LC3A and LC3C. Recombinant spartin-UBR–HA was incubated with recombinant GST, GST–LC3A, GST–LC3B or GST–LC3C, followed by GST pulldown and detected with anti-GST and anti-HA antibodies. c, SUM159 cells lacking spartin, transiently expressing LAMP1–mNG and mScarlet-I–spartin full-length (FL) or mScarlet-I–spartin–ΔUBR. Cells were treated with 0.5 mM OA for 24 h, then replaced to the complete medium 3 h before image acquisition. LDs were stained with BODIPY493/504. Scale bars: full-size, 20 μm; insets, 5 μm. d, Overlap of spartin FL or ΔUBR and LAMP1 in cell periphery, quantified by Pearson’s coefficient analysis. Mean ± s.d., n = 10 cells from three independent experiments, ****P < 0.0001, two-tailed unpaired t-test. e, AlphaFold2-based ColabFold structural prediction of the interaction between the UBR domain of spartin and LC3A. f, Recombinant spartin–UBR–HA or spartin-UBR ΔLIR (deletion of residues 193–200)–HA was incubated with recombinant GST or GST–LC3A, followed by GST pulldown and detected with anti-GST and anti-HA antibodies. g, Confocal imaging of live SUM159 cells lacking spartin, transiently expressing EGFP–LC3A and mScarlet-I–spartin FL or mScarlet-I–spartin–ΔLIR. Cells were treated with 0.5 mM OA for 24 h, then replaced to the complete medium 3 h before image acquisition. Scale bars: full-size, 20 μm; insets, 2 μm. h, Overlap between spartin FL or ΔLIR and LC3A in cell periphery shown in g was quantified by Pearson’s coefficient analysis. Mean ± s.d., n = 16 cells (Spartin FL) and 15 cells (ΔLIR) from three independent experiments, ****P < 0.0001, two-tailed unpaired t-test. Source numerical data and unprocessed blots are available in source data.