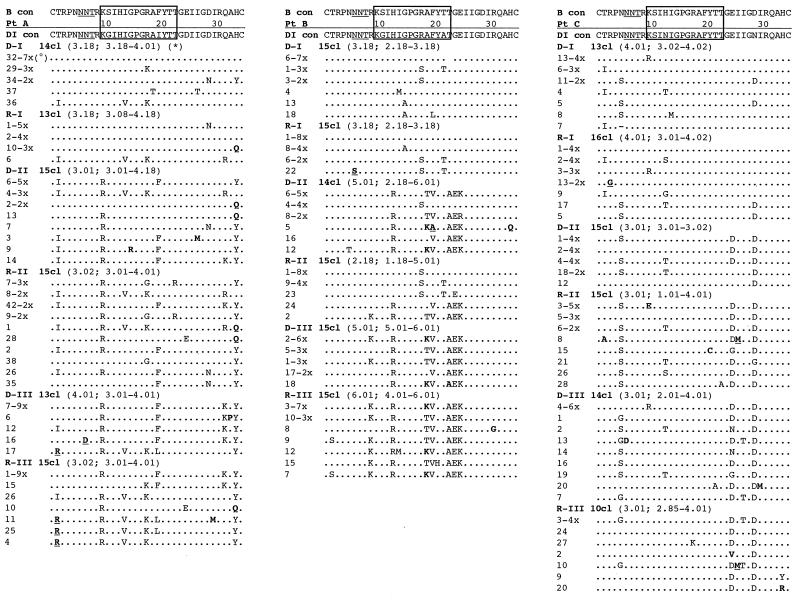
FIG. 5.
Deduced amino acid sequence alignments of the V3 loop from the six subjects. The B-clade consensus sequence reported above each alignment shows the most common amino acid found in each position among 1,078 viral variants (see reference 49). The sequences from each subject are aligned with the majority consensus sequence from the first proviral sample (DI con) at the top of each alignment. The clinical sample from which the sequences were derived, i.e., proviral DNA or plasma RNA, are indicated as D or R, respectively. The serial time points are indicated by Roman numbers, and the actual number of clones sequenced within each sample is indicated by a number followed by “cl” (clones). The deduced amino acid sequences are identified by the clone number. Dots indicate identity with the reference sequence, while dashes represent gaps introduced to maintain the alignment. Underlined residues indicate unique variants not identified before, and residues in boldface indicate vary rare variants (<0.5%). The box at the top of each alignment identifies the principal neutralization domain; the N-linked glycosylation site is underlined. Symbols: *, median net charge and range at physiological pH; °, frequency of clones with identical amino acid sequences.