Figure 1.
Global suppression of Tnfsf11 using CRISPRi causes osteopetrosis
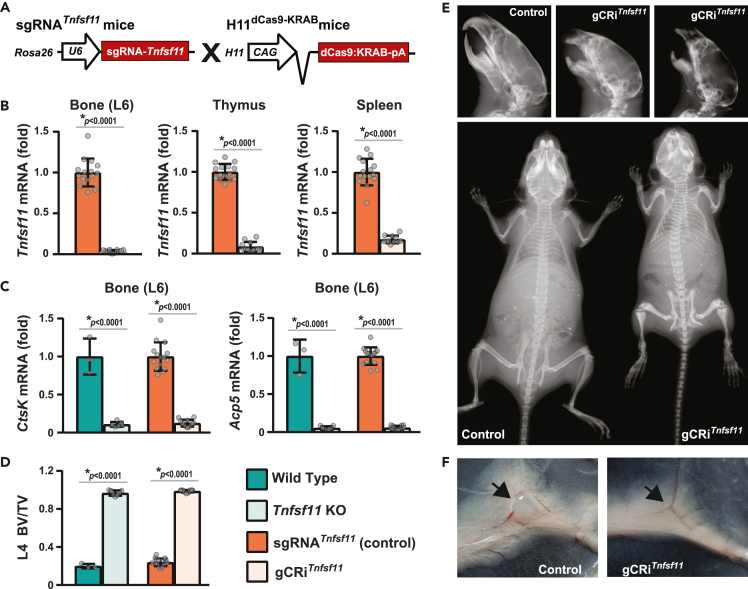
(A) sgRNATnfsf11 mice were crossed with H11dCas9-KRAB mice to globally suppress Tnfsf11.
(B–F) Gene expression and skeletal analysis of Tnfsf11 knock-out mice (Tnfsf11 KO, n = 7), their wild type littermates (n = 3); H11dCas9-KRAB; sgRNATnfsf11 mice (gCRiTnfsf11, n = 12), and their littermate controls (sgRNATnfsf11 or Control, n = 15) were performed at 5 weeks of age. Both sexes were included. B, Tnfsf11 mRNA levels were measured in the bone (lumbar vertebrae 6, L6), thymus, and spleen of gCRiTnfsf11 mice and their littermate controls by quantitative real-time PCR (qRT-PCR). ∗p < 0.0001 as calculated by unpaired t-test with Welch’s correction (L6 and spleen), or by Rank-Sum test (thymus). Data presented as mean ± standard deviation (SD). C, CtsK and Acp5 mRNA levels were measured in bones of Tnfsf11 KO, gCRiTnfsf11 mice, and their littermate controls by qRT-PCR. For all qRT-PCR analyses, mRNA levels were normalized to mouse Actb and indicated as fold (normalized to the mean of the corresponding littermate control values). D, Vertebral cancellous bone volume over tissue volume (BV/TV) of Tnfsf11 KO mice, gCRiTnfsf11 mice and their littermate controls were measured by μCT analysis of lumbar vertebra 4 (L4). C and D, Data presented as mean ± SD, ∗p < 0.0001 comparing Tnfsf11 KO or gCRiTnfsf11 to their corresponding littermates; #p < 0.05 comparing wild type to sgRNA, or Tnfsf11 KO to gCRiTnfsf11 as assessed by one-way ANOVA with Tukey adjustment. E, Skull and whole body X-ray images were generated at five weeks of age. F, Arrows point to the presence or lack of inguinal lymph nodes in the control and gCRiTnfsf11 mouse images. Data presented as mean + SD.