Abstract
A detailed metabolomic study was performed on various maturation stages of Murraya koenigii fruit pulps, seed, and leaf. Among the fruit pulps, stage 6 had the highest TPC (13.27 mg/g of GAE) and TFC content (6.16 mg/g RE). The extracts also showed promising free radical scavenging activity, especially in the seed (IC50DPPH 427 μg/mL). Metabolomics study revealed the identification of 133 metabolites in fruit pulps, seeds and leaves using the METLIN database. In silico PASS software analysis predicted the antimutagenic property of myricetin and bismurrayaquinone A. Pathway analysis revealed the phenylpropanoid biosynthesis pathway as one of the major pathways present in the fruit pulps. This detailed metabolic report of M. koenigii fruit maturation report brings a new insight into phytochemicals and their distribution in seed, pulps and leaves along with nutritive values and can be considered for nutritive and therapeutic purposes.
Keywords: Maturation stages, Targeted and non-targeted metabolomics, UHPLC-QTOF-IMS, METLIN database
1. Introduction
Plants have a rich history of being used as medicinal herbs for disease prevention and in foods for centuries. For nearly 1000 years, many plant species have been identified as potential sources for creating therapeutic medicines [1]. The majority of pharmaceuticals used today are natural compounds produced by plants Table S1. Numerous edible fruits are used as food sources and medicine because they offer substantial nutritional advantages, therapeutic effects and importantly their taste and flavour. Phenolic compounds are categorized into classes based on their structural properties: phenolic acids, flavanones, flavones, flavan-3-ols, anthocyanins, terpenoids etc. The primary food sources of phenolic compounds are fruits which contain phenolic acids, flavonoids, stilbenes, and lignins. Flavonoids are also important antioxidants that contribute greatly to free radical scavenging [2] Strong evidence of phenolic compounds being beneficial to human health, particularly linked to their antioxidant qualities, has drawn a lot of attention in recent decades [3]. According to published research, fruit Phenolics can enhance metabolic indicators often linked to several non-communicable disorders, including diabetes, hypertension, and obesity [4]. Fruit ripening is a genetically regulated process that involves variations in physiology, biochemistry, molecular structure, metabolites and flavouring [5]. Fruit's maturity stages may have an impact on the content of metabolites. There are a number of changes to the fruit's chemical, physical, and biological qualities as it ages. The amount and makeup of several metabolites alter the fruit's quality characteristics noticeably during the development and ripening phases. The main determinants of fruit quality are flavour, colour, and nutrient content [6]. This study's combination of metabolome and transcriptome analysis revealed substantial links between sugars, acids, flavonoid metabolites, and associated genes, indicating that these metabolites may be essential to fruit growth and development and ripening. The occurrence of metabolite breakdown mechanisms during the fruit maturation process altered the nutritional content of the fruit, making it intriguing to assess metabolites in connection to the fruit ripening stage [7]. Understanding the metabolomic alterations during the various fruit ripening phases also involves non-targeted metabolomics. Gallego et al., 2022, used a non-targeted metabolomic approach to characterise metabolic profiles associated with the ideal cacao harvest period in their study on the non-targeted metabolomics of Theobroma cacao fruit [8]. Similarly, mapping of metabolomic changes in Luffa aegyptiaca fruits during different maturation stages was performed using MS-based metabolomics to understand the biological changes during maturity [9].
Many studies over the years have attempted to understand the biochemistry and regulation of specific metabolic pathways, but it has only been in the last two decades, with the development and optimization of new technologies and the availability of Omics platforms, that it has been possible to understand the metabolic changes of plants [10]. Fruit metabolomic profiling is one of the trending technologies that has been proven to be a very important tool having numerous applications such as identifying fruit metabolites [11], variation in metabolites during stress conditions, maturation stages, metabolite variation during storage and so on. Studying the maturity stages and parts of fruit gives information about the metabolite up-regulation and down-regulation, metabolic pathway formation during different stages and also distribution of metabolites in different parts of the fruit [12].
Murraya koenigii Spreng. (L.) is a spice plant native to the Indian subcontinent belonging to the Rutaceae family. It is mostly cultivated in the tropics for the leaves' therapeutic benefits and also for its culinary uses [13,14]. It is also known as the Indian curry leaf plant, curry bush and curry leaf tree [15]. It has been used in various forms for centuries and is revered in Indian Ayurveda as "krishnanimba" [16]. A rising section of the worldwide population today highlights the value of herbal medicine. Previously antioxidant activity, antimicrobial activity, the prebiotic potential of M. koenigii and the anticancer activity of M. koenigii extracts for its fermented beverage was reported [17,18]. Wild fruits present in the Himalayan region are underutilized because of a lack of knowledge about the fruit and its therapeutic advantages, previously detailed metabolomic studies of underutilized fruit Cordia myxa (Assyrian plum) from the Himalayan region reported [19]. Similarly, metabolomics of another Indian spice hing (Ferula assa-foetida) was reported by Singh et al., 2023 [20]. Although M. koenigii leaves are widely used in culinary practices and therapeutic applications, the fruits on the other hand fruits are underutilized and also there are very few scientific works of literature about the fruit despite having various beneficial properties [21]. Metabolic changes during fruit maturation stages of M. koenigii have not been previously reported. Therefore, the current investigation has attempted qualitative and quantitative metabolomics of various maturation stages of M. koenigii fruit pulps including seed and leaf. The present study will be useful to determine and understand the metabolomic changes in the M. koenigii fruit during its maturation stages. Variations in the nutritive value of the fruit pulps, phenolic and flavonoid levels amongst all developmental stages have been successfully analyzed. Furthermore, using an advanced metabolomics approach major biosynthesis pathways midst the maturity stages have been investigated while identifying metabolites of interest with future biological active properties. According to current publications, curry leaves are heavily emphasized for their health benefits, whereas fruits receive less attention. This study will offer a thorough description of M. koenigii fruit, which will entice phytochemists and plant researchers to do more research into health applications.
2. Materials and methods
2.1. Chemicals and reagents
The chemicals used were of analytical reagent grade and HPLC grade. Methanol and DPPH (2,2-diphenyl-1-picrylhydrazyl) were obtained from Sigma-Aldrich, India. The standard working solutions of phenolic acid compounds (trans-cinnamic acid, chlorogenic acid, gallic acid, trans-ferulic acid, vanillic acid, p-coumaric acid, caffeic acid) and standard flavonoids (apigenin, myricetin, catechin, quercetin, rutin) were obtained from Sigma-Aldrich, India Pvt. Ltd. Sodium carbonate, aluminium chloride, sodium nitrite, Folin-Ciocâlteu reagent, sodium hydroxide, ascorbic acid and ABTS (2,2’-azino-bis 3-ethylbenzothiazoline-6-sulphonic acid) was obtained by Himedia, Pvt. Ltd.
2.2. Plant material
Six growth stages of fruits of M. koenigii (Stage I-Green, Stage II-Light green, Stage III-Light pink, Stage IV-Pink, Stage V-Purple, and Stage VI-Dark purple) (Fig. 1 A) were harvested along with leaves in the month of July 2022 from Palampur area, Himachal Pradesh, India (32.1109° N,76.5363° E). The fruits and leaves were refrigerated at −20 °C for further use.
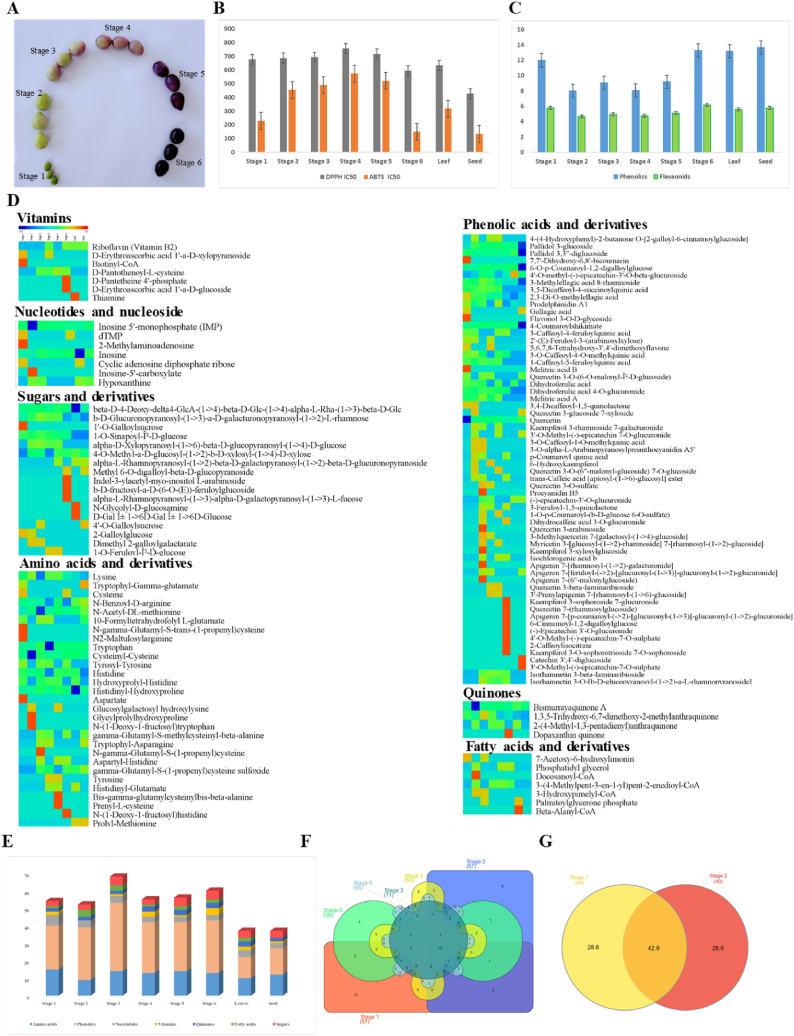
Fig. 1.
A. Different maturity stages of M. koenigii fruits B. In vitro antioxidant activity M. koenigii fruit pulps, seed and leaf C. TPC and TFC M. koenigii fruit pulps, seed and leaf D. Heatmaps of various metabolites found across all stages, leaf and seed of M. koenigii based on up-regulation and down-regulation of metabolites. Red indicates the up-regulation of metabolites and blue indicates the down-regulation of metabolites E. Number of metabolites present in all pulp stages including leaf and seed F. Venn diagram representing common metabolites between all pulp stages G. Venn diagram representing common metabolites between leaf and seed.
2.3. Proximate assay
Proximate composition analysis was performed for moisture, crude fibre, fat, crude protein, ash content and carbohydrates [22].
2.4. Extract preparation
Six different growth stages of M. koenigii were selected based on the colour and size pattern. Fruit pulps were separated in all six stages based on the size and colour of the fruits and the stages of seeds are not considered as it is not fully developed properly and can't be easily separated from the fruit. While in stage 6 seed was considered for providing an overview of phytochemicals in the seed. Fresh pulp (1 g), seed (1 g) and leaf (1 g) were macerated using mortar and pestle and transferred to a centrifuge tube containing 4 mL 70% methanol which was vortexed for 15 min and centrifuged at 7000 rpm, 10 min at 4 °C (Remi India Pvt. Ltd). The extractions were repeated twice with 3 mL 70% methanol making a final extract volume of 10 mL, filtered through a 0.22 μm filter and the filtrate was employed for further analysis.
2.5. Total phenolics, flavonoid content and in vitro antioxidant activity assay
Each M. koenigii extract was subjected to total phenolics and flavonoid estimation as previously reported [23,24]. The in vitro antioxidant activity of the extracts using DPPH and ABTS was estimated as previously reported [25,26].
2.6. Metabolomic profiling using UHPLC-QTOF-IMS
2.6.1. Targeted metabolomics
A gradient approach using ultra-high performance liquid chromatography quadrupole time of flight ion mobility mass spectrometry (UHPLC-QTOF-IMS) (Agilent Technologies, USA) was employed for the quantification and identification of phenolic acids and flavonoids [27]. Comparative standards included trans-cinnamic acid, rutin, gallic acid, quercetin, myricetin, trans-ferulic acid, catechin, caffeic acid, vanillic acid, and apigenin. Additionally, calibrations of every single standard were used for quantification. Phenolics and flavonoids present in the fruit pulps, seed and leaf skin were quantified and compared using a UHPLC-QTOF-IMS gradient method. Using column Eclipse Plus C18 RRHD (2.1 mm 150 mm, 1.8 m), the phenolic components of the extract were isolated. In the continuous gradient system, the mobile phases were water (A) and acetonitrile (B), each containing 0.1% and 0.05% formic acid, respectively. By changing the ratio of mobile phases, A and B, various gradient elution techniques were used to isolate the required phenolics. To achieve the exact separation of phenolics, several gradient combinations—0–2 min, 95% A; 2–6 min, 85% A; 6–9 min, 65% A; 9–13 min, 55% A; 17–20 min, 80% A; 20–22 min, and 95% A—were optimized. The injection volume range of 0.5–2 μL at a flow rate of 0.1–0.2 mL/min was set. Chromatograms were analyzed at 280 nm. Three copies of each processed sample were examined for statistical significance [28].
2.6.2. Non-targeted metabolomics and data processing
Extracts from all stages were subjected to non-targeted metabolomics, using a 6560-ion mobility quadrupole time-of-flight (IMQ-TOF) combined with LC-MS, (Agilent Technologies, USA). In a prior work gradient and mass spectrometry parameters were described [28]. The MassHunter Workstation Programme was used to assess the samples (Agilent Technologies, A02.02). Raw data (.d) were loaded into the Mass Hunter Qualitative Analysis programme (v. B.06.00, Agilent technology) to produce total fragments. Based on their m/z ratio, retention times, and ion intensities, extracted metabolites were transferred from total ion chromatograms (TICs) using the molecular feature extraction (MFE) approach. However, the precise mass and retention time thresholds were 0.10 min and 15 ppm, respectively [28,29].
2.7. Statistical analysis
Studies that included quantifying and determining content were carried out in triplicate (n = 3), and findings are presented as mean ± standard deviation (SD). Microsoft Excel and the statistics programmes used to create bar graphs. The Multiple Experiment Viewer (MeV, version 4.6.2) and interactive Venn 3 software were used to create heat maps and Venn diagrams. To give a comprehensive picture of the intrinsic variance of metabolites detected in various maturation stages including leaves and seeds, principal component analysis (PCA) was performed. PCA, PLS-DA, sPLS-DA, and pathway analysis were produced using MetaboAnalyst (version 5.0).
2.8. Biological activity prediction using PASS software
The biological activity of metabolites detected at greater concentrations in M. koenigii extracts was determined using the Prediction of Activity Spectra for Substances (PASS) software [30]. This in silico tool can show the likely activity or probable activity (Pa) and likely inactivity or probable inactivity (Pi) among various bioactive properties. The existing range of Pa and Pi values is 0–1.0, and in general, Pa, =Pi = 1 determined the likely values separately. Values starting at Pa >0.07 suggested a higher likelihood of biological activity. Thus, only anticipated bioactivity values greater than 0.07 were taken into account in the current study.
3. Results and discussion
3.1. Proximate analysis
The current study measured and analyzed the distribution of nutritional components in M. koenigii pulp, seed, and leaf. As the fruit matures, moisture levels were found to drop at different stages of development, leaves and seeds had less moisture than fruit pulps at various stages. Protein concentration decreased as the fruit became mature and was highest in the leaf (8.04%) and seed (4.8%). In fruit pulps, protein concentration varied from 1.62 to 3.80%, with stage 1 pulp having the highest content (3.89%) and stage 6 pulp having the lowest (1.62%). Ash content in M. koenigii decreased as the fruit matured (stage 1> stage 2 > stage 3> stage 4> Leaf > stage 5 > stage 6 > seed). Fibre content also showed decreasing trend as the fruit matured, its content ranged from 0.3 to 1.84% with the highest being stage 6 pulp, leaf and seed had a higher quantity of fibre with 6.96% and 3.8% respectively. Fat content was high in the initial stages but decreased during the final stages. The fat content of fruit pulps ranged from 1.51 to 6.43% with the lowest being stage 6 pulp. From the nutritional and consumption perspective leaf extract had a balanced nutritional profile compared to other pulp and seed extracts with the highest amount of crude fibre and protein content. However, carbohydrate content was higher in seed extract, this might be because sugars play an important role in seed during the germination process. In the case of fruit pulps stage 6 pulp had relatively higher crude fibre content and carbohydrate content. Stage 1 pulp also had higher protein and ash content which was at the highest levels compared to the other extracts indicating the presence of high mineral content. Proximate analysis of fruit maturation stages and seed have not been previously reported but leaf proximate values showed a similar correlation with another study [17,18]. The detailed proximate values are represented in Table 1.
Table 1.
Proximate analysis of M. koenigii pulp of different maturity stages, leaf and seed.
| Samples | Moisture % | Fat % | Crude fiber % | Crude protein % | Ash % | Carbohydrates % |
|---|---|---|---|---|---|---|
| Stage 1 | 81.71 ± 0.43a | 3.47 ± 0.07c | 0.30 ± 0.18e | 3.89 ± 0.04c | 9.03 ± 0.06a | 1.67 ± 0.14e |
| Stage 2 | 81.92 ± 0.06a | 3.35 ± 0.03c | 0.39 ± 0.16e | 3.87 ± 0.05c | 9.06 ± 0.06a | 1.39 ± 0.13e |
| Stage 3 | 80.16 ± 0.04a | 6.43 ± 0.03a | 1.02 ± 0.04d | 2.32 ± 0.21d | 8.84 ± 0.12a | 1.21 ± 0.21e |
| Stage 4 | 76.55 ± 0.03b | 5.35 ± 0.05b | 1.13 ± 0.08d | 2.16 ± 0.03d | 8.74 ± 0.03a | 6.05 ± 0.07d |
| Stage 5 | 75.90 ± 0.03b | 2.58 ± 0.03d | 1.65 ± 0.09c | 2.06 ± 0.05e | 8.65 ± 0.04a | 9.15 ± 0.16c |
| Stage 6 | 74.55 ± 0.04c | 1.51 ± 0.05e | 1.84 ± 0.04c | 1.62 ± 0.07f | 6.82 ± 0.03b | 13.64 ± 0.15b |
| Leaf | 55.81 ± 1.84d | 5.42 ± 0.29b | 6.96 ± 0.16a | 8.04 ± 0.12a | 8.78 ± 0.59a | 14.97 ± 1.63b |
| Seed | 43.98 ± 0.86e | 2.36 ± 0.04d | 3.28 ± 0.15b | 4.80 ± 0.08b | 3.85 ± 0.11c | 41.70 ± 0.67a |
Data are expressed as mean values (average of three replicates n = 3); Mean values with different superscripts within each column indicate statistically significant (p < 0.05 Tukey test).
3.2. Total phenolic content (TPC) and total flavonoid content (TFC) and invitro antioxidant activity
Fresh extracts of different fruit maturation stages of M. koenigii along with seed and leaf were investigated for their phenolic and flavonoid content. TPC in pulps of different maturity stages ranged from 8.03 to 13.65 mg GAE/g with the lowest content in stage 3 pulp and the highest being in stage 6 pulp. Different maturation stages of fruit pulps showed a parabolic trend with stage 1 pulp having 12.00 ± 0.16 mg GAE/g and stage 6 pulp having 13.65 ± 0.17 mg GAE/g, leaf and seed was found to have 13.19 ± 0.2 mg GAE/g and 13.65 ± 0.17 mg GAE/g respectively. This might be because of the downregulation of phenolic acid biosynthesis during the intermediate stages where the fruit is greenish in colour. During the final stages fruit gains deep purple colour which can be attributed to the increase in phenolic acid biosynthesis, hence the fruit shows a parabolic trend in TPC concentration. Flavonoids are also important antioxidants that contribute greatly to free radical scavenging. TFC content in different fruit maturation stages of pulps ranged from 4.67 to 6.16 mg RE/g with the highest present in stage 6 pulp and the lowest in stage 2 pulp. Similarly, leaf and seed also had a high amount of TFC with 5.57 ± 0.13 mg RE/g and 5.8 ± 0.11 mg RE/g, respectively. The in vitro antioxidant potential of pulps, seed and leaf was assessed by DPPH and ABTS assay and the results were reported as IC50. Seed extract showed the highest in vitro antioxidant activity compared to pulps and leaf extracts. In vitro antioxidant activity of the extracts in increasing order (Seed > stage 1> stage 6> leaf > stage 2> stage 3>stage 4>stage 5). The current results can be correlated with previous studies [18,31] which used extracts from different parts of the plant like leaf and bark. The detailed results are represented in Table 2, Fig. 1 B & 1 C.
Table 2.
TPC, TFC, DPPH, and ABTS of M. koenigii pulp of different maturity stages, leaf and seed.
| Samples | TPC (mg GAE/g of sample) | TFC (mg RE/g of sample) | DPPH IC50 in μg | ABTS IC50 in μg |
|---|---|---|---|---|
| Stage 1 | 12.00 ± 0.16c | 5.78 ± 0.16b | 678.16b | 229.39d |
| Stage 2 | 9.09 ± 0.15d | 4.67 ± 0.14e | 687.17b | 454.83b |
| Stage 3 | 8.03 ± 0.18e | 4.95 ± 0.20d | 694.07b | 489.57b |
| Stage 4 | 8.04 ± 0.19e | 4.77 ± 0.12e | 757.06a | 574.21a |
| Stage 5 | 9.19 ± 0.11d | 5.13 ± 0.1d | 717.40a | 521.76a |
| Stage 6 | 13.27 ± 0.25b | 6.16 ± 0.13a | 596.23d | 149.95e |
| Leaf | 13.19 ± 0.26b | 5.57 ± 0.13c | 633.98c | 318.03c |
| Seed | 13.65 ± 0.17a | 5.8 ± 0.11b | 427.18e | 134.52e |
| Vitamin C | NA | NA | 8.80 | 2.87 |
Data are expressed as mean values (average of three replicates n = 3); Mean values with different superscripts within each column indicate statistically significant (p < 0.05 Tukey test), NA: Not applicable.
3.3. Targeted metabolomics of phenolics and flavonoids
Eight known phenolic acids and flavonoids such as rutin, myricetin and catechin chlorogenic acid, caffeic acid, and p-coumaric acid were evaluated in the extracts. All fruit pulp maturation stages, including the leaf and seed contained Chlorogenic acid, one of the prevalent phenolic compounds, its content ranged from 74.59 to 320.57 μg/g, with the highest concentration in stage 6 pulp. Myricetin one of the major metabolites in M. koenigii [32], was also present in all the extracts with the highest being in the stage 6 pulp (411.09 μg/g) and the lowest being in leaves (40.71 μg/g). Catechin a flavonoid was found to be in the initial maturation stages of the fruit pulps and seed with the highest being in stage 1 (2163.10 μg/g). Ferulic acid is known to have a cardioprotective effect and is commonly present in fruits [33] ranging from 72.74 to 229.87 μg/g with the highest content in stage 6 pulp and lowest in stage 4 pulp. In a previous study, transcriptomics of M. koenigii leaves was performed and the study indicated out of all the secondary metabolism flavonoids biosynthesis was about 4.3% and phenylpropanoid synthesis was about 19.1% [34]. Comparing seed and leaf, the chlorogenic content was high in the leaf (164.91 μg/g) and relatively lower in the seed (112.45 μg/g). Only chlorogenic acid and myricetin were common to both the seed and leaf. Overall, during the initial maturation stages pulps had a higher concentration of phenolic compound, except stage 6 pulp. The variation in the phenolic compound might be because of water supply reduction and other environmental stress conditions. In a previous report impact of water supply reduction on phenolic compounds from mango pulp and peel was studied, the study showed increased phenolic compounds during reduced water supply to the plant [35]. Targeted metabolite concentration showed a parabolic trend in fruit pulp maturity stages. All samples were analyzed in triplicates (p ≤ 0.05) to achieve statistical significance. A detailed list of compounds with their concentration and chromatograms has been represented in Table 3 and Figure S2.
Table 3.
UHPLC-QTOF-IMS based quantification of phenolic acids and flavonoids in M. koenigii extracts.
| Phenolic compounds (μg/g) | Stage 1 | Stage 2 | Stage 3 | Stage 4 | Stage 5 | Stage 6 | Leaf | Seed |
|---|---|---|---|---|---|---|---|---|
| Chlorogenic acid | 242.03 ± 2.00b | 95.70 ± 12.57e | 121.59 ± 3.21d | 74.59 ± 1.80f | 87.93 ± 4.83e | 320.57 ± 2.54a | 164.91 ± 9.94c | 112.45 ± 5.54d |
| Catechin | 2163.10 ± 12.43a | NQ | 118.14 ± 5.50b | 89.08 ± 15.20c |
NQ | 89.50 ± 7.26c | NQ | 110.56 ± 13.95b |
| Rutin | NQ | 643.28 ± 20.60b | 495.04 ± 2.80c | 393.38 ± 16.54d | 657.75 ± 20.55b | 724.04 ± 11.54a | NQ | 173.93 ± 22.18e |
| Caffeic acid | 18.14 ± 2.04a | NQ | NQ | NQ | NQ | NQ | NQ | NQ |
| Vanillic acid | 23.73 ± 0.65b | NQ | NQ | NQ | NQ | 32.190 ± 2.99a | 24.56 ± 0.97b | NQ |
| Coumaric acid | 43.11 ± 0.73c | 9.75 ± 1.09e | 58.16 ± 1.50b | 24.40 ± 0.54d | 9.18 ± 1.65e | 24.467 ± 1.61d | 119.30 ± 1.33a | NQ |
| Ferulic acid | 73.78 ± 2.93e | 130.99 ± 3.56c |
167.89 ± 4.39b | 72.74 ± 1.72e | 27.272 ± 3.32f | 88.67 ± 2.67d | 229.87 ± 2.41a | NQ |
| Myricetin | 123.99 ± 1.90g | 248.9 ± 5.60d | 386.68 ± 8.0b | 266.57 ± 2.50c | 190.65 ± 2.72e | 411.09 ± 7.55a | 40.71 ± 4.32h | 142.28 ± 2.99f |
Data are expressed as mean values (average of three replicates n = 3); Mean values with different superscripts within each row indicate statistically significant (p < 0.05 Tukey test), NQ: Not quantifiable.
3.4. Metabolite profiling using UHPLC-QTOF-IMS
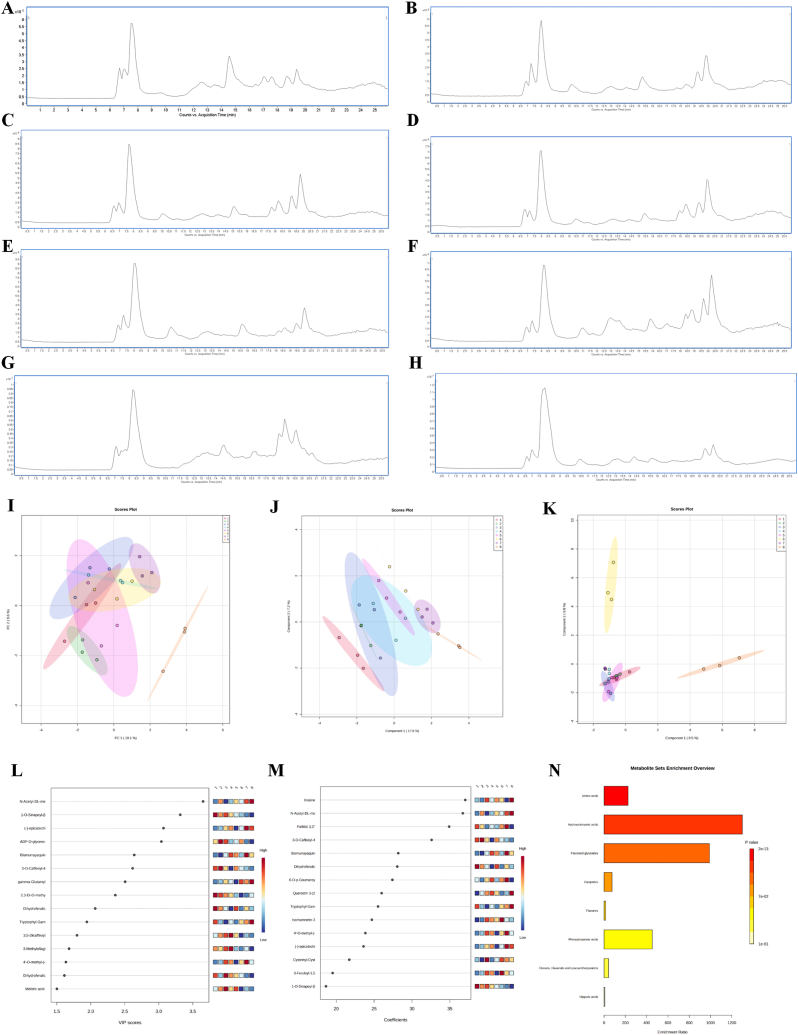
A total of 19 peaks were identified in all eight extracts of M. koenigii based on retention time (RT), comparison of molecular weight, and mass fragmentation pattern with literature (Table 4). Peaks were observed with good resolution in negative ionization mode. Out of the identified metabolites, one organic acid (Peak 5), one triterpene (Peak 14), seven were phenolic acid and its derivatives (Peaks 1, 2, 4, 6, 7, 10, 18), and ten were flavonoids and its derivatives (Peaks 3, 8, 9, 11, 12, 13, 15, 16, 17, 19). Metabolite profiling chromatograms have been shown in Figure S1.
Table 4.
Metabolites profiling using UHPLC-QTOF-IMS consisting of major fragments and expected compounds in M. koenigii fruit pulp maturation stages (1–6) respectively, (7) Leaf, (8) Seed.
| Peak No. | RT | Molecular formula | Theoretical mass | Measured mass | Major fragments | Expected compound | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | Metabolite Class |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1. | 7.516 | C9H8O4 | 180.02 | 179.03 [M − H]- | 132 | Caffeic acid | + | – | – | – | – | – | – | – | Phenolic acid |
| 2. | 7.98 | C15H17O9 | 342.30 | 341.11 [M − H]- | 179 | Caffeic acid hexoside | – | + | – | – | – | – | – | – | Phenolic acid glycoside |
| 3. | 10.103 | C15H14O6 | 290.26 | 289.90 [M − H]- | 207 | Catechin/epicatechin | – | – | + | + | + | + | + | + | Flavonoid |
| 4. | 10.933 | C13H16O10 | 332.26 | 331.90 [M − H]- | 301 | 1-O-galloyl hexose | – | – | – | – | – | + | – | – | Phenolic acid glycoside |
| 5. | 12.595 | C₆H₈O₇ | 192.03 | 191.02 [M − H]- | 170,179 | Citric acid | + | + | + | + | + | + | + | + | Organic acid |
| 6. | 12.968 | C16H20O9 | 356.32 | 355.06 [M − H]- | 191 | Ferulic acid hexoside | – | – | – | – | – | + | + | + | Phenolic acid glycoside |
| 7. | 13.649 | C16H17O9 | 354.02 | 353.09 [M − H]- | 179,191 | 3-caffeoylquinic acid | + | + | + | + | + | + | + | + | Phenolic acid |
| 8. | 14.121 | C21H18O13 | 478.4 | 477.96 [M − H]- | 451 | Quercetin 3-O-glucuronide | – | + | – | – | + | – | – | – | Flavonoid glycoside |
| 9. | 16.01 | C24H22O15 | 550.4 | 549.01 [M − H]- | 517 | Quercetin O-malonyl-O-hexoside | – | – | + | – | – | – | – | – | Flavonoid glycoside |
| 10. | 16.435 | C16H17O8 | 338.31 | 337.05 [M − H]- | 173,191 | 4-p-Coumaroyl quinic acid | + | – | – | – | – | – | – | – | Phenolic acid glycoside |
| 11. | 17.552 | C27H30O16 | 610.5 | 609.14 M − H]- | 463 | Quercetin 3-rutinoside | – | – | – | – | – | + | + | – | Flavonoid glycoside |
| 12. | 17.578 | C23H22O13 | 506.4 | 505.10 [M − H]- | 463 | Quercetin-3-O-acetyl hexoside | + | + | + | + | + | + | + | + | Flavonoid glycoside |
| 13. | 17.794 | C21H19O12- | 464.4 | 463.09 [M − H]- | 301,179 | Quercetin 3-O-hexoside | – | + | + | + | + | + | + | – | Flavonoid glycoside |
| 14. | 18.442 | C30H48O5 | 488.7 | 487.30 [M − H]- | 463 | Tormentic acid | – | + | – | + | – | – | – | – | Triterpene |
| 15. | 18.72 | C15H10O6 | 286.24 | 285.17 [M − H]- | 255 | Kaempferol | + | – | – | – | – | – | – | – | Flavonoid |
| 16. | 20.22 | C24H20O14 | 534.4 | 533.09 [M − H]- | 515,499 | Kaempferol-O-malonyl-O hexoside | + | + | + | + | + | + | + | + | Flavonoid glycoside |
| 17. | 21.08 | C20H18O11 | 434.3 | 433.23 [M − H]- | 301,178 | Querecetin-3-O-pentoside | + | – | – | – | – | – | – | – | Flavonoid glycoside |
| 18. | 24.087 | C15H17O8 | 326.30 | 325.18 [M − H]- | 119 | p-coumaric acid hexoside | – | + | – | – | + | – | – | – | Phenolic acid glycoside |
| 19. | 24.170 | C32H28O14 | 636.56 | 635.35 [M − H]- | 449 | Kaempferol-O-acetyl-O-p-coumaroyl-O-hexoside | – | + | – | – | – | – | – | – | Flavonoid glycoside |
3.4.1. Phenolic acid and derivatives
Phenolic acids like caffeic acid (Peak 1), caffeic acid hexoside (Peak 2), 1-O-galloyl hexose (Peak 4), ferulic acid hexoside (Peak 6), 3-caffeoylquinic acid (Peak 7), 4-p-coumaroylquinic acid (Peak 10), p-coumaric acid hexoside (Peak 18) were identified at RT 7.516, 7.98, 10.933, 12.968, 13.649, 16.435, 24.087 with a mass of 179.03 [M − H]-, 341.11 [M − H]-, 331.90 [M − H]-, 355.06 [M − H]-, 353.09 [M − H]-, 337.05 [M − H]-, 325.18 [M − H]- respectively.
3.4.2. Flavonoid derivatives and others
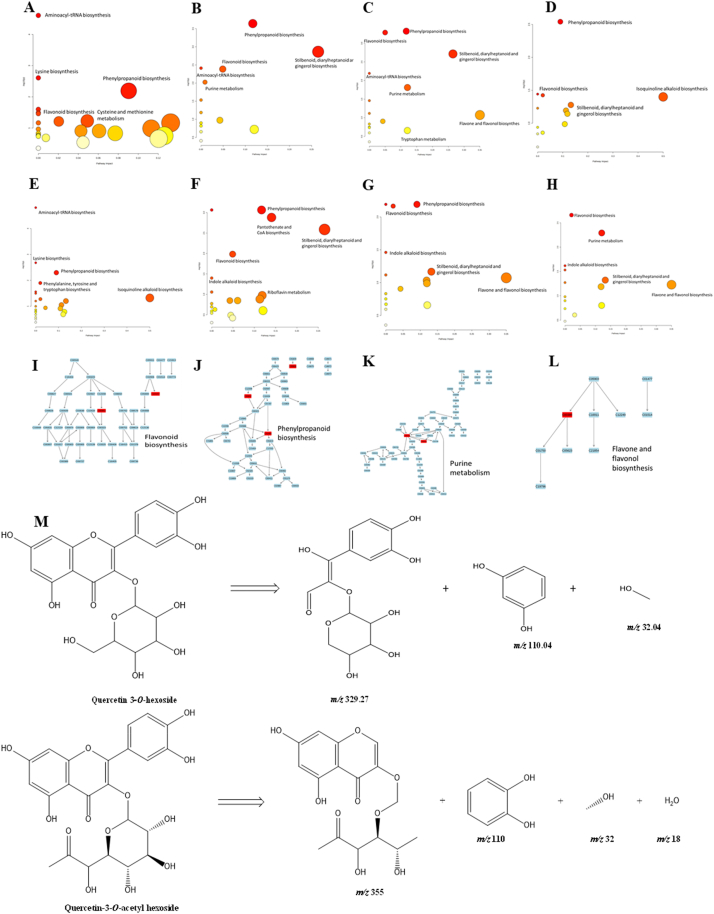
A common component of the human diet is flavonoids, a diverse family of natural chemicals that are extensively found as secondary metabolites in higher plants [36]. Ten flavonoid derivatives were identified in all the extracts. Catechin/epicatechin (Peak 3), quercetin 3-O-glucuronide (Peak 8), quercetin O-malonyl-O-hexoside (Peak 9), quercetin 3-rutinoside (Peak 11), quercetin-3-O-acetyl hexoside (Peak 12), quercetin 3-O-hexoside (Peak 13), kaempferol (Peak 15), kaempferol-O-malonyl-O hexoside (Peak 16), querecetin-3-O-pentoside (Peak 17), and kaempferol-O-acetyl-O-p-coumaroyl-O-hexoside (Peak 19) with RT 10.103, 14.121, 16.01, 17.552, 17.578, 17.794, 18.72, 20.22, 21.08, 24.170 and mass of 477.96 [M − H]-, 549.01 [M − H]-, 337.05 [M − H]-, 505.10 [M − H]-, 463.09 [M − H]-, 285.17 [M − H]-, 591.13 [M − H]-, 533.09 [M − H]-, 433.23 [M − H]-, 635.35 [M − H]- respectively were identified. Tormentic acid (Peak 14) and citric acid (Peak 5). Mass fragmentation of Quercetin-3-O-acetyl hexoside and Quercetin 3-O-hexoside are shown in Fig. 3 M.
Fig. 3.
A-H. Pathway analysis of stage 1 to stage 6 pulp, leaf and seed respectively I. Flavonoid biosynthesis J. Phenylpropanoid biosynthesis K. Purine metabolism L. Flavone and flavonoid biosynthesis M. Mass fragmentation pattern.
3.5. Non-targeted metabolomics
Metabolite identification using the METLIN database and visualization Raw (.d) files obtained from UHPLC-QTOF-IMS were used to determine the metabolites. The received files were searched against the METLIN database and 133 metabolites were identified among which 29 amino acids, 62 phenolic compounds, 7 nucleobases, 7 vitamins, 4, quinones, 7 fatty acids derivatives and 17 sugars and their derivatives were present. Out of all the stages, stage 3 pulp had the highest number of identified metabolites 68. Secondly in stage 6 pulp 60 metabolites were identified. Leaf and seed had the lowest number of identified metabolites of 37 each which indicates relatively lower activity of secondary metabolism in leaf and seed. (Fig. 1 E). A total of 15 fatty acids were identified in all six stages, leaf and seed out of which stage 2 pulp and stage 3 pulp had the highest number of identified fatty acids, this data can also be linked with the proximate fat content of stage 3 pulp which had highest fat of 6.43%. Heatmaps generate a matrix based on the higher and lower regulation of the metabolites present [37]. Metabolite’s relative up-regulation and down-regulation were represented in the form of heatmaps (Fig. 1 D), the red colour indicates the compound is down-regulated and blue indicates the up-regulation of compounds, among all the 6 maturity stages of pulps including leaf and seed, phenolic compounds were the major class of metabolites followed by amino acids and sugars. The distribution of metabolites in different fruit pulp maturity stages was visualised in the form of a Venn diagram (Fig. 1F–G). The highest number of metabolites were present in stage 3 pulp followed by stage 6 and stage 5 pulps. Out of 133 metabolites identified 31 metabolites were common to all the stages of the pulp mainly, lysine, pallidal 3-glucoside, prodelphinidin A1, 4-coumaroylshikimate and dihydroferulic acid 4-O-glucuronide were present in all the stages. PCA, PLS-DA, (Partial least squares-discriminant analysis) sPLS-DA (Sparse PLS discriminant analysis) was used to determine the variation between the metabolites during various maturity stages of the fruit [38]. PCA is one of the common chemometric techniques used to determine the variation between multiple datasets [39]. Metabolite variation between the fruit maturity stages, leaves and was were low according to the PCA plot with PC1 (18.1%) and PC 2 (9.6%). PC 1 and PC 2 accounted for 27.7% variation between metabolites of different fruit pulp stages, seed and leaf. Which indicates there is a high similarity between the stages. However, the metabolites present in the seed showed high variation from that of other clusters in the datasets and had clear separation on the plot. The leaf metabolite cluster was also separated from the rest of the clusters. The stage 2 metabolites also had slight metabolite variation. Fruit pulp stages 4,5 and 6 and the highest similarities can be observed in (Fig. 2 I). Overall, there were similarities between datasets of different fruit pulp stages which indicates similar metabolomic makeup of fruit pulps. The identified metabolites were subjected to PLS-DA. For classification purposes, biomarker selection in metabolomics investigations, and variable importance in projection (VIP) value, partial least squares-discriminant analysis (PLS-DA) is utilized [40]. VIP value can describe the weight of the independent variable in explaining the dependent variable. The score plot showed low variability between the metabolites, represented by C1 (17.6% variance) and C2 (7.2% variance). The plot also represents considerable variation in the seed which is grouped away from the pulps and leaf (Fig. 2 J). Sparse PLS Discriminant Analysis (sPLS-DA) is used to categorise the samples, the sparse variant allows the determination of the best predictive or discriminative characteristics in the data [41]. Again, the sPLS-DA showed low variation between the stages except for the seed and stage 6 pulp. The plot showed (8.5% C1) and (9.8% C2) and stages 1 to stage 5 pulp metabolites formed a low variance cluster whereas stage 6 pulp and seed were highly dispersed from the cluster which shows the difference in the metabolite’s formation in seed and stage 6 pulp compared to other stages and leaf (Fig. 2 K). VIP score projection and coefficient analysis were performed on metabolites to determine the different metabolite fold changes between the stages. Metabolites are considered differentially expressed if the VIP value is more than 1 and the p-value is less than 0.05 and the VIP value can describe the weight of the independent variable in explaining the dependent variable [42]. Fifteen different metabolites were found as a result of the comparison, with VIP >1, FDR less than 0.05, and FC > 1 as the criteria. Among the metabolites, 9 were phenolic compounds, 3 belonged to amino acids and 3 were of other classes. Dihydroferulic acid, 2,3-Di-O-methyl ellagic acid, N-Acetyl-DL-methionine exhibited major reduction. VIP score and coefficient analysis were represented in (Fig. 2 L-M, Table S2 & S3).
Fig. 2.
A-H. Indicates MS spectra of stage 1 to stage 6 pulp, leaf and seed respectively I. PCA score plots J. PLS-DA score plot K. sPLS-DA score plot L. VIP score projections M. VIP coefficient N. Pathway enrichment analysis of all the metabolites present in every stage of pulp including seed and leaf.
3.6. Analysis of metabolic pathways and interactions
Pathway enrichment studies of all the metabolites present in every stage, seed and leaf of M. koenigiie extracts were performed using MetaboAnalyst 5.0 to determine the metabolites belonging to different classifications and distribution. Among the classifications hydroxycinnamic acids, flavonoid glycosides and polypropionic acids had higher P values whereas amino acids, dipeptides and flavones had low P values which indicate this class of metabolites is down-regulated in fruit pulps (Fig. 3 N) Pathway studies were utilized to compare the metabolite formation in the maturity stages of the fruit to understand the distribution of metabolites and to determine the biological role in the metabolic pathway. The Biosynthesis of M. koenigii fruit metabolite at different maturity levels was also studied using pathway studies (MetaboAnalyst, version 5.0). Metabolites that were identified using the METLIN database were incorporated into the column labelled compound list, MetaboAnalyst software was used to generate pathway analysis of compounds. The results obtained were based on the upward and down-ward regulation of metabolites and their intensity. The significance cut-off was at a p-value of 0.05 to determine the metabolite trend in the fruits. The major metabolic pathway involving identified metabolites were represented in Figure (3 A-H). According to the results obtained, phenylpropanoid biosynthesis and aminoacyl-tRNA biosynthesis were the major pathways in the stage 1 fruit pulp. As the fruit matured biosynthesis of aminoacyl-tRNA was gradually reduced. Phenylpropanoid biosynthesis, flavonoid biosynthesis, stilbenoid, diarylheptanoid and gingerol biosynthesis were three major biosynthesis pathways common to all M. koenigii fruit stages including the leaves. A higher number of metabolic pathways were seen in stage 1 pulp and stage 6 pulp, these two stages contain a higher number of metabolites compared to other stages. Phenylpropanoids are a diverse class of secondary metabolites that originate from the amino acids phenylalanine or tyrosine. They undergo various enzymatic reactions to form different groups of compounds, such as flavonoids, monolignols, phenolic acids, stilbenes, and coumarins. These groups have distinct chemical structures and biological functions in plants and animals [43]. Light induces phenylpropanoid biosynthesis according to a previous report which indicates that there are increased secondary metabolites if the plants are grown in the presence of light, similarly, synthesis of light-inducible and light-independent anthocyanins was reported different genes are responsible for regulating light-inducible and light-independent anthocyanins [44,45]. Metabolites identified in this pathway are cis-2-hydroxycinnamate 2-coumarinate (C05838), chlorogenic acid (C00852) and ferulic acid (C01494) (Fig. 3 J). This pathway further gives rise to other secondary metabolite pathways such as flavonoid biosynthesis, coumarins, stilbenes and catechin pathways. The flavonoid biosynthesis pathway is a part of phenylpropanoid synthesis giving rise to mostly of flavonoids [46,47]. Flavonoids are compounds with a 15C structure that has two aromatic rings (A and B) and a pyrane ring (C). The C6–C3–C6 skeleton can vary in saturation, substitution and oxidation of the pyrane ring, leading to different flavonoid classes. Some examples are flavones (apigenin, luteolin), flavonols (kaempferol, quercetin, myricetin), flavanones (naringenin, hesperitin), flavanonols (dihydrokaempferol, dihydroquercetin), isoflavones (rotenone), aurones (sulphuretin), anthocyanidins (cyanidin), and proanthocyanidins. Flavones are formed by the chalcones specifically naringenin chalcone which is converted naringenin which is further converted to dihydroflavonol like dihydrokaempferol. Naringenin also gives rise to flavones. Dihydrokaempferol further gives rise to flavonols, dihydroquercetin and dihydromyricetin. This compound gives to anthocyanins through a series of enzymatic reactions [48]. Metabolites identified in this pathway are myricetin (C10107) (Fig. 3 I). Other pathways in the fruit pulps were purine metabolism with identified metabolites like IMP (C00130) and inosine (C00294) (Fig. 3 K) and flavone and flavonol biosynthesis with quercetin (C00389) (Fig. 3 L). Some of the minor pathways identified were isoquinoline alkaloid biosynthesis, pantothenate and CoA biosynthesis, lysine biosynthesis and tryptophan metabolism (Table S4).
3.7. Bioactive property prediction using in silico PASS software
Myricetin is a flavonoid that is a major compound of M. koenigii and chlorogenic acid which is present in all the maturity stages of the fruits including seeds and leaves identified by targeted metabolomics. Bismurrayaquinone A, a member of carbazoles, prodelphinidin A1 an anthocyanin and pallidol 3-glucoside are present in most of the stages of the fruit pulps. These five compounds were subjected to in silico PASS software. According to previous reports, the PASS programme is an effective in silico method for predicting the bioactive properties of molecules [49]. Therefore, by entering the specifics of detected compounds from M. koenigii, biological activities were predicted utilising this computer algorithm. The results were obtained by clicking on the Mol file while uploading the compound's structures which were downloaded from the PubChem database. A table with likely activity (Pa) and unlikely inactivity (Pi) values for a range of biological activities ranging from 0 to 1.0 was obtained by choosing the predict button once. To get the highest possible biological activity that might be taken into consideration for future experimental models, values were further fixed at Pa 0.7. The results obtained from the software exhibited that myricetin has (0.963 Pa) for antimutagenic and (0.924 Pa) for antioxidant properties. Myricetin has widespread cytotoxic effects, and its mechanisms are unique [50] Similarly, chlorogenic Acid (0.856 Pa) for free radical scavenging activity and (0.846 Pa) for anticarcinogenic. Bismurrayaquinone A had (0.844 Pa) for antineoplastic effect. Prodelphinidin A1 had a Pa of 0.766 for antimutagenic properties. Pallidol 3-glucoside showed (0.836 Pa) for anticarcinogenic properties, and all the selected metabolites showed Pa <0.7, indicating a high probability of their biological property. This information can be used for future experimental studies.
4. Conclusion
M. koenigii is an underutilized fruit found in the Indian subcontinent. The consumption of this fruit and its health benefits is not very well understood, and it has little or no scientific evidence. The current study has shown that the fruit has phytochemicals and nutrients, which can be attributed to nutritive value and health-promoting factors. TPC and TFC of the fruit pulps of different maturity stages, seed and leaf were performed and found that there was a parabolic trend in the phenolic and flavonoid concentration as the fruit matured. Targeted metabolomics of the different maturation stages of pulp revealed various phytochemicals, changes in their concentration during maturation stages, and the presence of phytochemicals in seed and leaf. Non-targeted metabolomics showed metabolite variation in different maturity stages and metabolite distribution. Heatmap represented the up-regulation and down-regulation of metabolites in different stages and pathway analysis showed the metabolites involved in different metabolic pathways. In vitro antioxidant activity was performed through ABTS and DPPH assays and the result showed considerable inhibition concentration. Finally, this study shows that M. koenigii fruits have nutritional, antioxidant properties due to the presence of phenolics and can be considered for edible and culinary purposes.
Author contribution statement
Manoj S Aroor, Vikas Dadwal: Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Robin Joshi: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Mahesh Gupta: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Data availability statement
Data included in article/supplementary material/referenced in article.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
We sincerely thank the Director of the CSIR-Institute of Himalayan Bioresource Technology for their support during the course of the project. For financial assistance, the authors further recognize the Council of Scientific and Industrial Research (CSIR), Project: HCP-0035. (Institutional manuscript ID No. 5229).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.heliyon.2023.e18688.
Contributor Information
Robin Joshi, Email: robinsjoshi@gmail.com, robinjoshi@ihbt.res.in.
Mahesh Gupta, Email: mgupta@ihbt.res.in.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Petrovska B.B. Historical review of medicinal plants’ usage. Phcog. Rev. 2012;6(11):1. doi: 10.4103/0973-7847.95849. 10.4103%2F0973-7847.95849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aljerf L., Aljerf N. Food products quality and nutrition in relation to public. Balancing health and disease. Prog. Nutr. 2023;25(N. 1) doi: 10.23751/pn.v25i1.13928. [DOI] [Google Scholar]
- 3.Zou Z., Xi W., Hu Y., Nie C., Zhou Z. Antioxidant activity of Citrus fruits. Food Chem. 2016;196:885–896. doi: 10.1016/j.foodchem.2015.09.072. [DOI] [PubMed] [Google Scholar]
- 4.Lima M.D.C., de Sousa C.P., Fernandez-Prada C., Harel J., Dubreuil J.D., De Souza E.L. A review of the current evidence of fruit phenolic compounds as potential antimicrobials against pathogenic bacteria. Microb. Pathog. 2019;130:259–270. doi: 10.1016/j.micpath.2019.03.025. [DOI] [PubMed] [Google Scholar]
- 5.Prasanna V., Prabha T.N., Tharanathan R.N. Fruit ripening phenomena–an overview. Crit. Rev. Food Sci. Nutr. 2007;47(1):1–19. doi: 10.1080/10408390600976841. [DOI] [PubMed] [Google Scholar]
- 6.Gong C., Diao W., Zhu H., Umer M.J., Zhao S., He N., Lu X., Yuan P., Anees M., Yang D., Kaseb M.O. Metabolome and transcriptome integration reveals insights into flavor formation of ‘Crimson’watermelon flesh during fruit development. Front. Plant Sci. 2021;12 doi: 10.3389/fpls.2021.629361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mubarok S., Yulianto F., Budiarto R., Rahmat B.P.N., Khoerunnisa S.A. Metabolite correlation with antioxidant activity in different fruit maturation stages of Physalis peruviana. Biodiversitas J. Biol. Diversity. 2021;22(5) doi: 10.13057/biodiv/d220536. [DOI] [Google Scholar]
- 8.Gallego A.M., Zambrano R.A., Zuluaga M., Rodríguez A.V.C., Cortés M.S.C., Vergel A.P.R., Valencia J.W.A. Analysis of fruit ripening in Theobroma cacao pod husk based on untargeted metabolomics. Phytochemistry. 2022;203 doi: 10.1016/j.phytochem.2022.113412. [DOI] [PubMed] [Google Scholar]
- 9.Maamoun A.A., El-Akkad R.H., Farag M.A. Mapping metabolome changes in Luffa aegyptiaca Mill fruits at different maturation stages via MS-based metabolomics and chemometrics. J. Adv. Res. 2021;29:179–189. doi: 10.1016/j.jare.2019.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nadella K.D., Marla S.S., Kumar P.A. Metabolomics in agriculture. OMICS A J. Integr. Biol. 2012;16(4):149–159. doi: 10.1089/omi.2011.0067. [DOI] [PubMed] [Google Scholar]
- 11.Dadwal V., Joshi R., Gupta M. Comparative metabolomics of Himalayan crab apple (Malus baccata) with commercially utilized apple (Malus domestica) using UHPLC-QTOF-IMS coupled with multivariate analysis. Food Chem. 2023;402 doi: 10.1016/j.foodchem.2022.134529. [DOI] [PubMed] [Google Scholar]
- 12.Mamat S.F., Azizan K.A., Baharum S.N., Noor N.M., Aizat W.M. GC-MS and LC-MS analyses reveal the distribution of primary and secondary metabolites in mangosteen (Garcinia mangostana Linn.) fruit during ripening. Sci. Hortic. 2020;262 doi: 10.1016/j.scienta.2019.109004. [DOI] [Google Scholar]
- 13.Adebajo A.C., Ayoola O.F., Iwalewa E.O., Akindahunsi A.A., Omisore N.O.A., Adewunmi C.O., Adenowo T.K. Anti-trichomonal, biochemical and toxicological activities of methanolic extract and some carbazole alkaloids isolated from the leaves of Murraya koenigii growing in Nigeria. Phytomedicine. 2006;13(4):246–254. doi: 10.1016/j.phymed.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 14.Iman V., Mohan S., Abdelwahab S.I., Karimian H., Nordin N., Fadaeinasab M., Noordin M.I., Noor S.M. Anticancer and anti-inflammatory activities of girinimbine isolated from Murraya koenigii. Drug Des. Dev. Ther. 2016:103–121. doi: 10.2147/DDDT.S115135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walde S.G., Jyothirmayi T., Rao P.P., Shivaswamy R., Srinivas P. Flavour volatiles of leaves, fruits and seed cotyledons of Murraya koenigii L. Flavour Fragrance J. 2005;20(2):169–172. doi: 10.1002/ffj.1436. [DOI] [Google Scholar]
- 16.Samanta S.K., Kandimalla R., Gogoi B., Dutta K.N., Choudhury P., Deb P.K., Devi R., Pal B.C., Talukdar N.C. Phytochemical portfolio and anticancer activity of Murraya koenigii and its primary active component, mahanine. Pharmacol. Res. 2018;129:227–236. doi: 10.1016/j.phrs.2017.11.024. [DOI] [PubMed] [Google Scholar]
- 17.Bhatt S., Dadwal V., Padwad Y., Gupta M. Study of physicochemical, nutritional, and anticancer activity of Murraya Koenigii extract for its fermented beverage. J. Food Process. Preserv. 2022;46(1) doi: 10.1111/jfpp.16137. [DOI] [Google Scholar]
- 18.Abeysinghe D.T., Kumara K.A.H., Kaushalya K.A.D., Chandrika U.G., Alwis D.D.D.H. Phytochemical screening, total polyphenol, flavonoid content, in vitro antioxidant and antibacterial activities of Sri Lankan varieties of Murraya koenigii and Micromelum minutum leaves. Heliyon. 2021;7(7) doi: 10.1016/j.heliyon.2021.e07449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar A., Joshi R., Vasishth A., Guleria V., Kumar D. UHPLC-QTOF-IMS-based metabolite fingerprinting of underutilized Cordia myxa fruits and leaves: a nutraceutical source. ACS Food Sci. Technol. 2022;2(5):793–807. doi: 10.1021/acsfoodscitech.1c00398. [DOI] [Google Scholar]
- 20.Singh P.P., Joshi R., Kumar R., Kumar A., Sharma U. Comparative phytochemical analysis of Ferula assa-foetida with Ferula jaeschkeana and commercial oleo-gum resins using GC-MS and UHPLC-PDA-QTOF-IMS. Food Res. Int. 2022 doi: 10.1016/j.foodres.2022.112434. [DOI] [PubMed] [Google Scholar]
- 21.Balakrishnan R., Vijayraja D., Jo S.H., Ganesan P., Su-Kim I., Choi D.K. Medicinal profile, phytochemistry, and pharmacological activities of Murraya koenigii and its primary bioactive compounds. Antioxidants. 2020;9(2):101. doi: 10.3390/antiox9020101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.AOAC M. Association of official analytical chemists. Official methods of analysis. AOAC: Off Methods Anal. 1990;1:69–90. [Google Scholar]
- 23.Ainsworth E.A., Gillespie K.M. Estimation of total phenolic content and other oxidation substrates in plant tissues using Folin–Ciocalteu reagent. Nat. Protoc. 2007;2(4):875–877. doi: 10.1038/nprot.2007.102. [DOI] [PubMed] [Google Scholar]
- 24.Kumari R., Gupta M. Elucidating the techno-functional, morphological and phenolic properties of hull less barley and buckwheat incorporated pasta. Food Chem. Adv. 2022;1 doi: 10.1016/j.focha.2022.100055. [DOI] [Google Scholar]
- 25.Sharma O.P., Bhat T.K. DPPH antioxidant assay revisited. Food Chem. 2009;113(4):1202–1205. doi: 10.1016/j.foodchem.2008.08.008. [DOI] [Google Scholar]
- 26.Joshi R., Rana A., Gulati A. Studies on quality of orthodox teas made from anthocyanin-rich tea clones growing in Kangra valley, India. Food Chem. 2015;176:357–366. doi: 10.1016/j.foodchem.2014.12.067. [DOI] [PubMed] [Google Scholar]
- 27.Dadwal V., Joshi R., Gupta M. A comparative metabolomic investigation in fruit sections of Citrus medica L. and Citrus maxima L. detecting potential bioactive metabolites using UHPLC-QTOF-IMS. Food Res. Int. 2022 doi: 10.1016/j.foodres.2022.111486. [DOI] [PubMed] [Google Scholar]
- 28.Dadwal V., Joshi R., Gupta M. A multidimensional UHPLC-DAD-QTOF-IMS gradient approach for qualitative and quantitative investigation of citrus and malus fruit phenolic extracts and edibles. ACS Food Sci. Technol. 2021;1(10):2006–2018. doi: 10.1021/acsfoodscitech.1c00323. [DOI] [Google Scholar]
- 29.Chong J., Soufan O., Li C., Caraus I., Li S., Bourque G., Wishart D.S., Xia J. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018;46(W1):W486–W494. doi: 10.1093/nar/gky310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siddiqui S., Upadhyay S., Ahmad I., Hussain A., Ahamed M. Cytotoxicity of Moringa oleifera fruits on human liver cancer and molecular docking analysis of bioactive constituents against caspase‐3 enzyme. J. Food Biochem. 2021;45(5) doi: 10.1111/jfbc.13720. [DOI] [PubMed] [Google Scholar]
- 31.Dadwal V., Bhatt S., Joshi R., Gupta M. Development and characterization of controlled released polyphenol rich micro‐encapsulate of Murraya koenigii bark extract. J. Food Process. Preserv. 2020;44(5) doi: 10.1111/jfpp.14438. [DOI] [Google Scholar]
- 32.Oliva E., Viteritti E., Fanti F., Eugelio F., Pepe A., Palmieri S., Sergi M., Compagnone D. Targeted and semi-untargeted determination of phenolic compounds in plant matrices by high performance liquid chromatography-tandem mass spectrometry. J. Chromatogr. A. 2021;1651 doi: 10.1016/j.chroma.2021.462315. [DOI] [PubMed] [Google Scholar]
- 33.Pandi A., Raghu M.H., Chandrashekar N., Kalappan V.M. Cardioprotective effects of Ferulic acid against various drugs and toxic agents. Beni-Suef Univ. J. Basic Appl. Sci. 2022;11(1):1–9. doi: 10.1186/s43088-022-00273-5. [DOI] [Google Scholar]
- 34.Meena S., Rajeev Kumar S., Dwivedi V., Kumar Singh A., Chanotiya C.S., Akhtar M.Q., Kumar K., Kumar Shasany A., Nagegowda D.A. Transcriptomic insight into terpenoid and carbazole alkaloid biosynthesis, and functional characterization of two terpene synthases in curry tree (Murraya koenigii) Sci. Rep. 2017;7(1) doi: 10.1038/srep44126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosalie R., Joas J., Mertz C., Dufossé L., Léchaudel M. Impact of water supply reduction and cold storage on phenolic compounds from mango (Mangifera indica L. cv. Cogshall) pulp and peel. Plants. 2022;11(22):3038. doi: 10.3390/plants11223038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin Y.S. Recent advances in natural antifungal flavonoids and their derivatives. Bioorg. Med. Chem. Lett. 2019;29(19) doi: 10.1016/j.bmcl.2019.07.048. [DOI] [PubMed] [Google Scholar]
- 37.Nkosi N.J., Shoko T., Manhivi V.E., Slabbert R.M., Sultanbawa Y., Sivakumar D. Metabolomic and chemometric profiles of ten southern African indigenous fruits. Food Chem. 2022;381 doi: 10.1016/j.foodchem.2022.132244. [DOI] [PubMed] [Google Scholar]
- 38.Pupo A., Ku K.M., Gallagher J.E. Effects of MCHM on yeast metabolism. PLoS One. 2019;14(10) doi: 10.1371/journal.pone.0223909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bakir S., Capanoglu E., Hall R.D., de Vos R.C. Variation in secondary metabolites in a unique set of tomato accessions collected in Turkey. Food Chem. 2020;317 doi: 10.1016/j.foodchem.2020.126406. [DOI] [PubMed] [Google Scholar]
- 40.Wu Q., Li Z., Chen X., Yun Z., Li T., Jiang Y. Comparative metabolites profiling of harvested papaya (Carica papaya L.) peel in response to chilling stress. J. Sci. Food Agric. 2019;99(15):6868–6881. doi: 10.1002/jsfa.9972. [DOI] [PubMed] [Google Scholar]
- 41.Medina-Meza I.G., VanderWeide J., Torres-Palacios C., Sabbatini P. Quantitative metabolomics unveils the impact of agricultural practices in the grape metabolome. ACS Agri. Sci. Technol. 2021;1(3):253–261. doi: 10.1021/acsagscitech.1c00043. [DOI] [Google Scholar]
- 42.Yi H., Yang G., Xiong Y., Wu Q., Xiao H., Wen X., Yang X., Wang L., Jiang Z. Integrated metabolomic and proteomics profiling reveals the promotion of Lactobacillus reuteri LR1 on amino acid metabolism in the gut–liver axis of weaned pigs. Food Funct. 2019;10(11):7387–7396. doi: 10.1039/C9FO01781J. [DOI] [PubMed] [Google Scholar]
- 43.Anwar M., Chen L., Xiao Y., Wu J., Zeng L., Li H., Wu Q., Hu Z. Recent advanced metabolic and genetic engineering of phenylpropanoid biosynthetic pathways. Int. J. Mol. Sci. 2021;22(17):9544. doi: 10.3390/ijms22179544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hemm M.R., Rider S.D., Ogas J., Murry D.J., Chapple C. Light induces phenylpropanoid metabolism in Arabidopsis roots. Plant J. 2004;38(5):765–778. doi: 10.1111/j.1365-313X.2004.02089.x. [DOI] [PubMed] [Google Scholar]
- 45.Ma Z.H., Li W.F., Mao J., Li W., Zuo C.W., Zhao X., Dawuda M.M., Shi X.Y., Chen B.H. Synthesis of light-inducible and light-independent anthocyanins regulated by specific genes in grape ‘Marselan’ (V. vinifera L.) PeerJ. 2019;7:e6521. doi: 10.7717/peerj.6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nabavi S.M., Šamec D., Tomczyk M., Milella L., Russo D., Habtemariam S., Suntar I., Rastrelli L., Daglia M., Xiao J., Giampieri F. Flavonoid biosynthetic pathways in plants: versatile targets for metabolic engineering. Biotechnol. Adv. 2020;38 doi: 10.1016/j.biotechadv.2018.11.005. [DOI] [PubMed] [Google Scholar]
- 47.Falcone Ferreyra M.L., Rius S.P., Casati P. Flavonoids: biosynthesis, biological functions, and biotechnological applications. Front. Plant Sci. 2012;3:222. doi: 10.3389/fpls.2012.00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu W., Feng Y., Yu S., Fan Z., Li X., Li J., Yin H. The flavonoid biosynthesis network in plants. Int. J. Mol. Sci. 2021;22(23) doi: 10.3390/ijms222312824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singh D., Gawande D.Y., Singh T., Poroikov V., Goel R.K. Revealing pharmacodynamics of medicinal plants using in silico approach: a case study with wet lab validation. Comput. Biol. Med. 2014;47:1–6. doi: 10.1016/j.compbiomed.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 50.Song X., Tan L., Wang M., Ren C., Guo C., Yang B., Ren Y., Cao Z., Li Y., Pei J. Myricetin: a review of the most recent research. Biomed. Pharmacother. 2021;134 doi: 10.1016/j.biopha.2020.111017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data included in article/supplementary material/referenced in article.