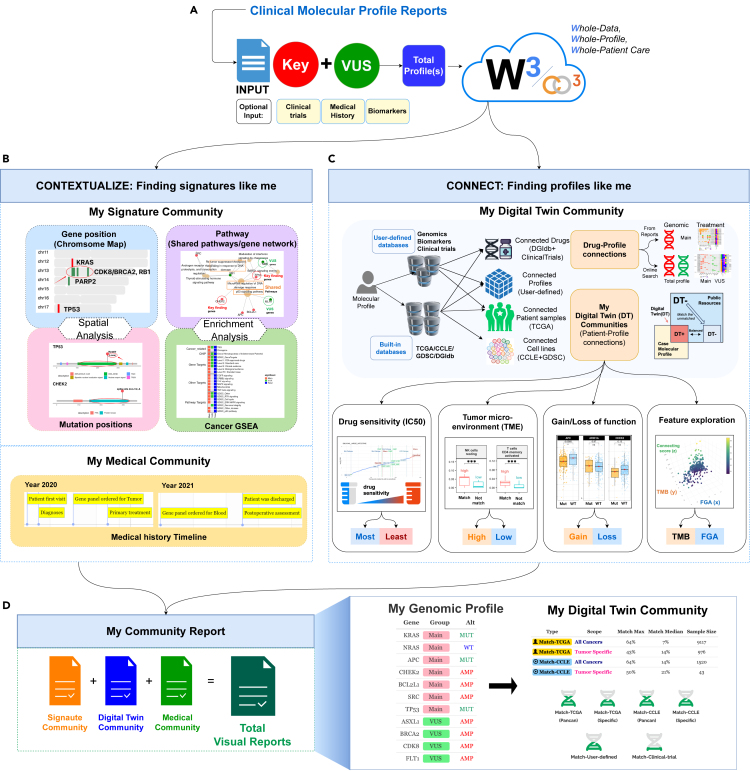
Figure 2.
Overview of W3 (whole-data, whole-profile, for whole-patient care) platform implementation of CO3 system design
(A) Input. Required input is a list of molecular alterations from two gene sets, key/main and variants of unknown significance (VUS) contained within a molecular profiling report. Optional input includes reported a list of clinical trials, a timeline of medical history of events (diagnosis, treatment, and testing), and biomarkers (e.g., TMB, MS Status, and PD-L1 expression)
(B) Context. This module finds (spatially and biologically enriched) ‘signatures like me’ using both the combined (total) and separate, key and VUS gene sets.
(C) Connect. This module finds ‘profiles like me’ by matching the total profile molecular alterations with public and user-defined data collections. Digital twin communities are determined based on user-defined, case profile matched (DT+) and unmatched (DT−) to public resources, with the option to further balance sample sizes between them. Once matched, several downstream analyses may be explored that include drug sensitivity testing from patient-derived cell lines, gain and loss of function prediction, the abundance of markers associated with the tumor microenvironment, and relationships among features.
(D) Report. An all-visual report based on analyses performed that includes a digital infographic summarizing the type of profile communities built and their degree of matching is output. See also Figures S1–S10.