Fig. 1.
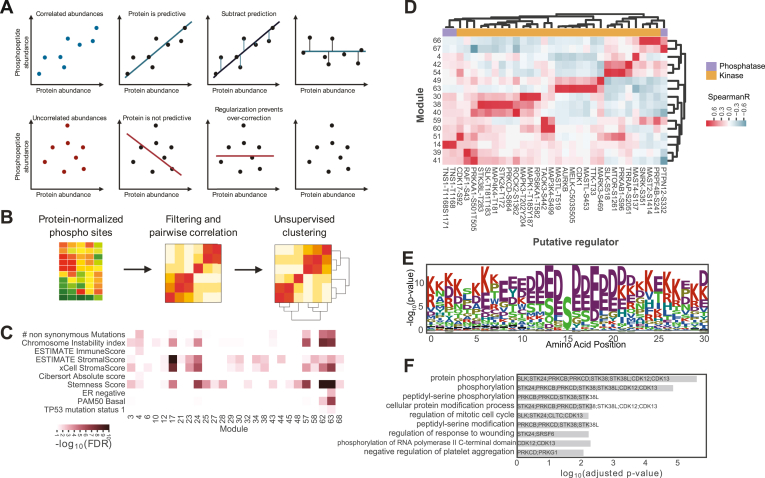
PhosphoDisco core functionalities.A, regularized linear models are used to normalize phosphorylation data by protein abundance. B, normalized phosphorylation data are correlated with itself and modules are found by clustering the correlation matrix using hypercluster. C, clinical metadata are correlated with module scores to find relevant modules. D, kinase and phosphatase abundances are correlated with module scores and assigned potential regulators to modules. E, motifs can be calculated from peptides for a module. F, enrichments of phosphosites in a module can be calculated against a phosphosite annotations database like PhosphositePlus.