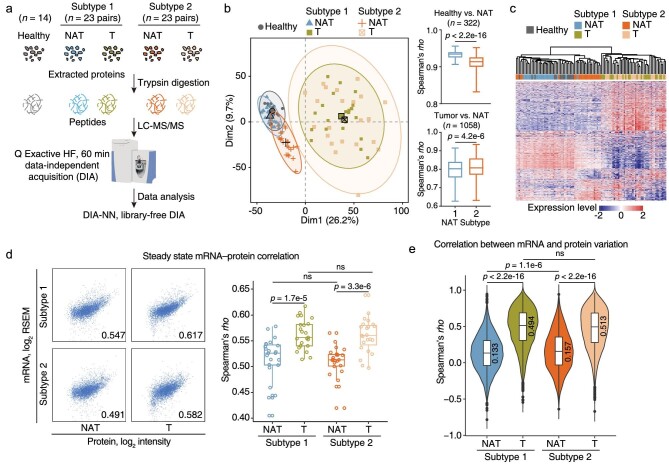
Figure 4.
Subtype 1 NATs were similar to healthy liver tissues. a. Workflow of data-independent acquisition (DIA)-based proteomic analysis of healthy livers (n = 14), Subtype 1 NATs and paired tumors (n = 23), and Subtype 2 NATs and paired tumors (n = 23). Proteins were extracted and subjected to trypsin digestion. The MS data were collected on a Q Exactive HF mass spectrometer. The DIA data were analyzed by DIA-NN software in a library-DIA mode. b. Left panel: PCA of healthy livers, Subtype 1 NATs and paired tumors, and Subtype 2 NATs and paired tumors. Right panel: pair-wise Spearman's correlation coefficients between NATs and healthy liver tissues (upper panel; n = 322, Mann–Whitney test); pair-wise Spearman's correlation coefficients between NATs and tumor tissues (lower panel; n = 1058, Mann–Whitney test). c. HCA of healthy livers, Subtype 1 NATs and paired tumors, and Subtype 2 NATs and paired tumors. d. Correlation of steady state mRNA-protein abundance. Left panel, representative scatter plots of mRNA-protein abundances of two NAT subtypes and their paired tumors. Right panel, the comparison between mRNA-protein correlation coefficients (rho) of NATs and that of tumors in each NAT subtype. Differences between NATs and their paired tumors were analyzed by Wilcoxon matched-pairs signed rank test. Differences between two NAT subtypes and differences between the paired tumors of two NAT subtypes were analyzed by Mann–Whitney test. e. Distribution of Spearman's correlation coefficients between mRNA and protein variation in two NAT subtypes and their paired tumors. The median coefficients are labeled above (Kolmogorov–Smirnov test).