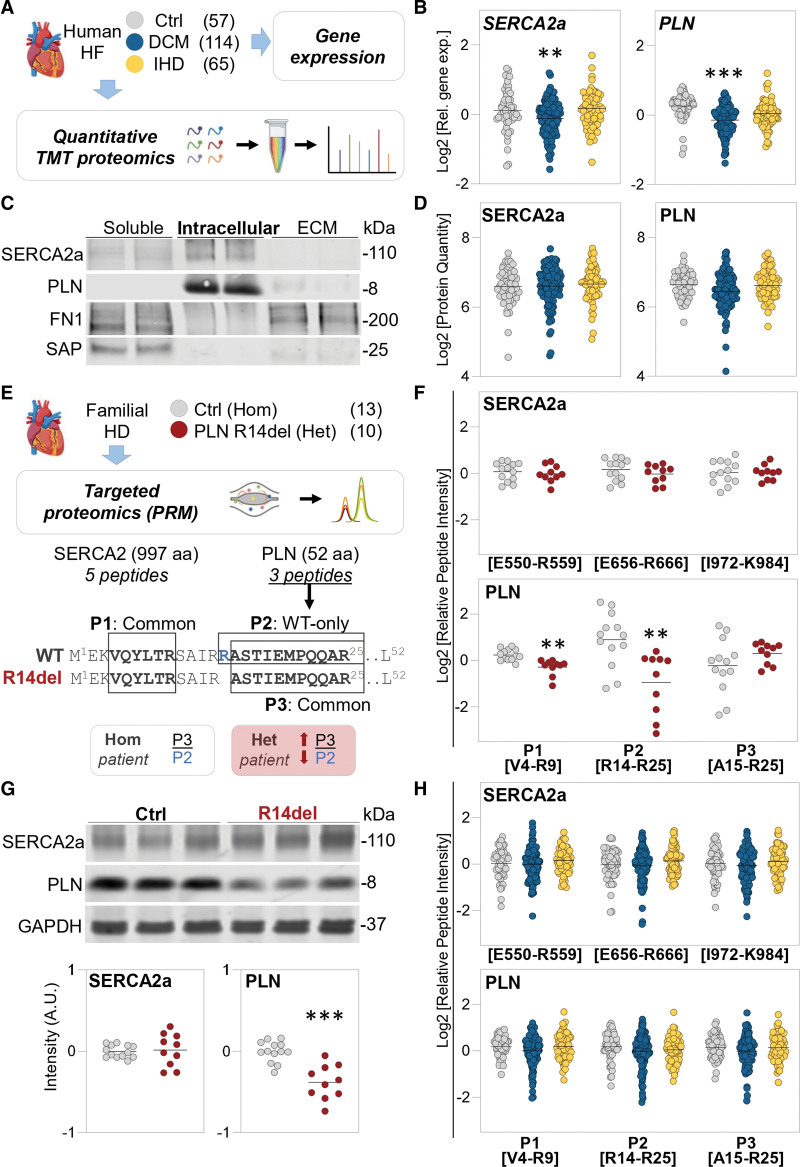
Figure.
SERCA2a protein levels in human heart failure. A, Schematic representation of the workflow used for analyzing cardiac explants from patients with end-stage heart failure (HF) and nonfailing hearts as controls (Ctrl). Gene expression was assessed by reverse transcription–quantitative polymerase chain reaction. The proteomic workflow includes tandem mass tag (TMT) labeling followed by mass spectrometry. B, Relative gene expression for sarcoplasmic reticulum Ca2+ATPase 2a (SERCA2a) and phospholamban (PLN). Black line represents the mean of the Log2 relative gene expression. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as normalizer. **P<0.01, ***P<0.001 by Ebayes empirical method of the limma package corrected for age and sex. C, Distribution of SERCA2a and PLN after a 3-step extraction process from cardiac tissue. A representative Western blot is shown with 2 pooled samples of the soluble, intracellular, and extracellular matrix (ECM)–enriched protein extracts. D, Cardiac protein levels of SERCA2a and PLN as measured by multiplexed TMT proteomics. Black line represents the mean of the Log2 protein levels, normalized and scaled to the sample pool in the TMT reference channel. No significant differences were detected by Ebayes empirical method of the limma package in both uncorrected analysis and after correction for age and sex. E, Schematic representation of the targeted proteomics workflow for analyzing hearts from patients with the R14del variation and nonfailing hearts as controls. Parallel reaction monitoring (PRM) was used to quantify 5 proteotypic peptides for SERCA2a and 3 proteotypic peptides for PLN. PLN peptide 1 (P1; V4–R9) and peptide 3 (P3; A15–R25) are common to both the wild-type (WT) and R14del alleles. PLN peptide 2 (P2; A14–R25) is WT specific. Trypsin cleaves PLN at either R13 or R14. The presence of the R14del variation shifts the tryptic cleavage site by 1 amino acid (from P2 to P3 in heterozygous [Het] patients). F, Quantification of SERCA2a and PLN by PRM in carriers of the R14del PLN variation and control hearts. Three representative peptide quantifications are shown for SERCA2a, and all 3 targeted peptide quantifications for PLN are displayed. The position of each peptide in the target protein is indicated. Black line represents the mean of the Log2 peptide intensity normalized to the total signal intensity per sample. **P<0.01 by Ebayes empirical method of the limma package and corrected for age and sex. G, Representative Western blot and relative quantification of SERCA2a and PLN on cardiac tissue lysates from patients with PLN carrying the R14Del variation and nonfailing hearts as controls. Intensity values are given as arbitrary units (A.U.) after normalization on GAPDH as loading control. ***P<0.001 by Ebayes empirical method of the limma package corrected for age and sex. H, SERCA2a and PLN protein levels were measured with PRM in explanted hearts from patients with end-stage HF and nonfailing hearts as controls. Three representative peptide quantifications are shown for SERCA2a. All 3 targeted peptide quantifications for PLN are shown. Each peptide position in the target protein is indicated. Black line represents the mean of the Log2 peptide intensity normalized to the total signal intensity per sample. No significant differences were detected by Ebayes empirical method of the limma package both in uncorrected analysis and after correction for age and sex. aa Indicates amino acid; DCM, dilated cardiomyopathy; FN1, fibronectin; HD, heart disease; Hom, homozygous; IHD, ischemic heart disease; and SAP, serum amyloid P component.