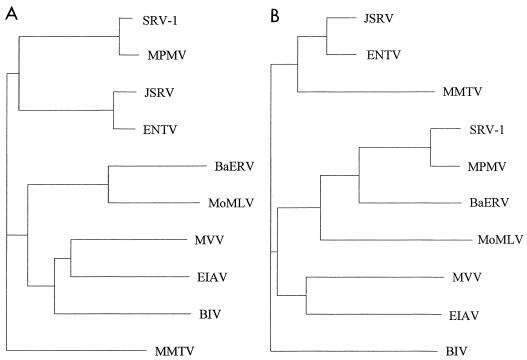
FIG. 5.
Phylogenetic trees of (A) gag and (B) env SU nucleotide sequences of ENTV, baboon endogenous retrovirus (BaERV) (18), bovine immunodeficiency virus (BIV) (11), equine infectious anemia virus (EIAV) (19), JSRV (38), MMTV (24), simian SRV-1 type D retrovirus (SRV-1) (27), MPMV (33), maedi-visna virus (MVV) (28), and Moloney murine leukemia virus (MoMLV) (32). The sequences were aligned by using GCG Eclustalw, and unrooted trees were constructed by the neighbor-joining method of Saitou and Nei (30) (Clustree). Horizontal branch lengths are proportional to evolutionary distances as estimated by percent divergence.