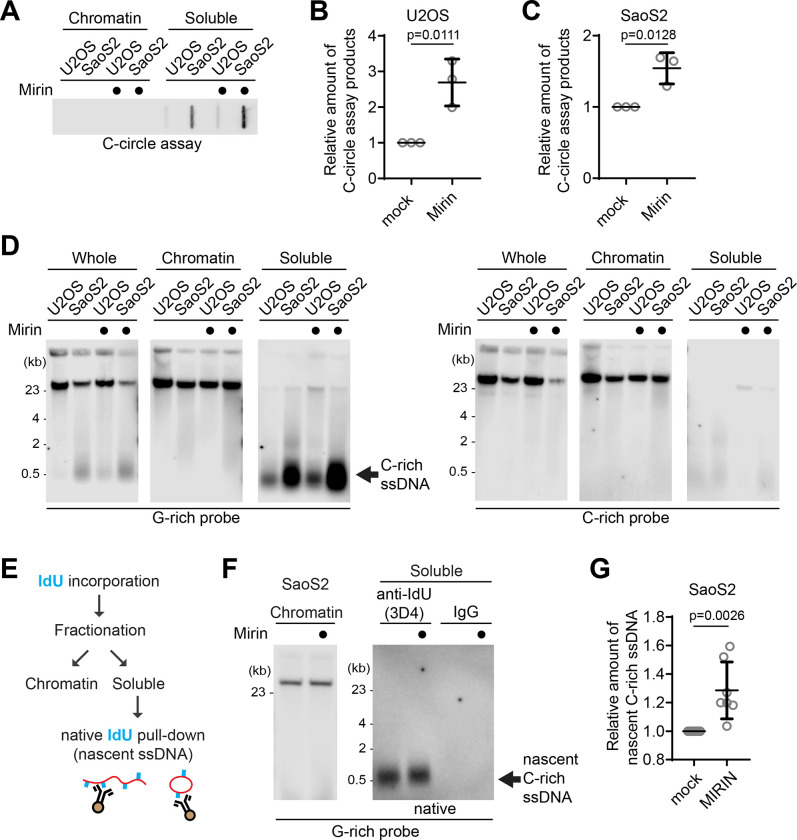
Figure 3. MRE11 nuclease inhibition leads to increases in C-circles and C-rich ssDNAs.
(A) C-circle assay for U2OS and SaoS2 cells with or without Mirin (MRE11 nuclease inhibitor) treatment. Samples were fractionated into chromatin and soluble fractions. DNA amount: 100ng. U2OS and Saos2 cells were treated with 50 μM Mirin for 48 hr. (B-C) Quantification of the C-circle assay in A; U2OS cells (B), and SaoS2 cells (C), as the relative amount of C-circle assay products (mean ± SD; unpaired t-test). (D) 4SET assay for U2OS and SaoS2 cells with or without Mirin treatment after fractionation of samples into whole, chromatin, and soluble fractions. DNA amount: 600 ng. G-rich probe was used to detect C-rich telomeric sequences, and C-rich probe was used to detect G-rich telomeric sequences. C-rich single stranded DNAs are indicated by an arrow. (E) Illustration depicting the process of pulling down nascent single-stranded DNA from the soluble fraction incorporated with IdU. (F) Native IdU pulldown assay using IdU (3D4) antibody for SaoS2 cells treated with either Mirin (50 μM Mirin and 100 μM IdU for 20 hours) or mock (100 μM IdU for 20 hours). IgG-negative control; chromatin DNA-control for quantity. (G) Quantification of the native IdU-pulldown assay in F; Relative amount of nascent C-rich ssDNAs (mean ± SD; unpaired t-test).