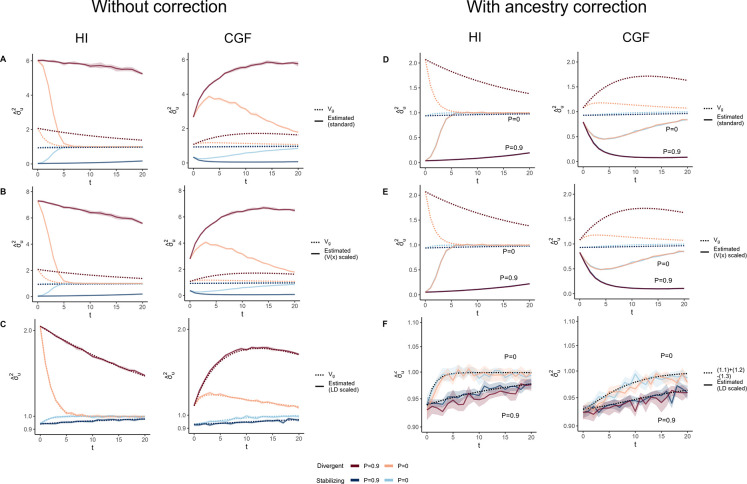
Figure 5:
Genetic variance estimated with HE regression in admixed populations under the HI (left column) and CGF (right column) models either without (A-C) or with (D-F) adjustment for individual ancestry. The solid lines represent estimates from simulated data averaged across ten replicates with red and blue colors representing estimates for traits under divergent and stabilizing selection, respectively. indicates the strength of assortative mating. (A & D) show the behavior of for the default scaling, (B, E) shows when the genotype at a locus is scaled by its sample variance ( scaled), and (C, F) when it is scaled by the sample covariance (LD scaled). The dotted lines in A-E represent the expected in the population based on Eq. 1 and in F, represent the expected after removing any genetic variance along the ancestry axis. The shaded areas represent the 95% bootstrapped confidence bands of the estimate.