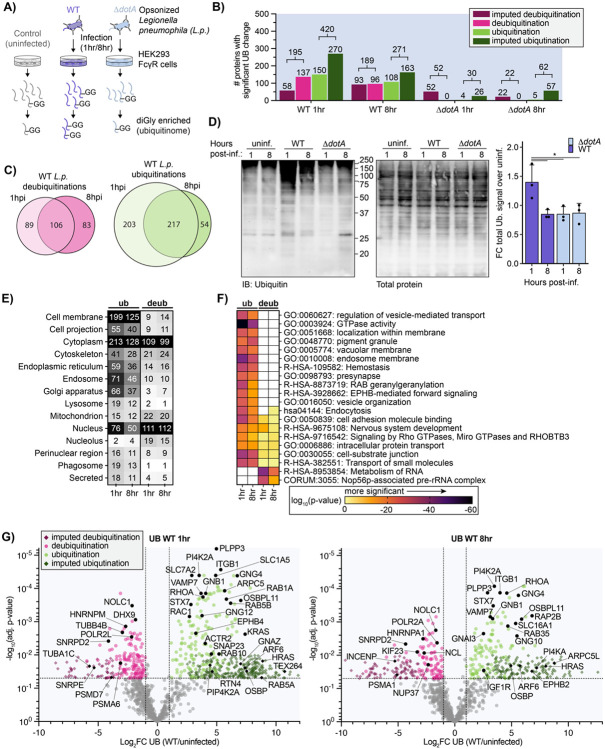
Figure 1: L.p. infection induces T4SS-dependent ubiquitinome changes in the host cell.
(A) Schematic of experimental procedures. (B) Counts of proteins with a significant increase (green) or decrease (magenta) in ubiquitination compared to uninfected control for the indicated infection conditions. (Significance threshold for all subsequent analysis: ∣log2(FC)∣>1, p<0.05, see note in text on use of imputation in this dataset). (C) Overlap of proteins with a significant increase (green) or decrease (magenta) in ubiquitination compared to uninfected control in the 1-hour vs 8-hour WT L.p. infected conditions. (D) Immunoblot analysis of the total pool of ubiquitinated proteins in HEK293T FcγR cells infected with WT or ΔdotA L.p. for 1 or 8 hours, or left uninfected. Invitrogen No-Stain protein labeling reagent was used to quantify total protein before immunoblot analysis. Total ubiquitin signal was first normalized to total protein for each sample, then the fold change over the appropriate uninfected sample was calculated. Data was subjected to a one-way ANOVA followed by a post-hoc Tukey-Kramer test for pairwise comparisons (* = p<0.05, n=3). (E) Subcellular localization analysis of proteins with a significant increase or decrease in ubiquitination compared to uninfected control during WT L.p. infection for 1 or 8 hours. (F) Pathway and protein complex analysis of proteins with a significant increase or decrease in ubiquitination compared to uninfected control during WT L.p. infection for 1 or 8 hours. Terms not significantly enriched for a given experimental condition are represented by white boxes. Analysis performed using Metascape (see methods). (G) Volcano plot representation of all ubiquitinome data in WT vs uninfected comparison at 1- and 8-hours post-infection. Imputed values are shown as diamonds. Significance threshold is indicated by the dotted line.