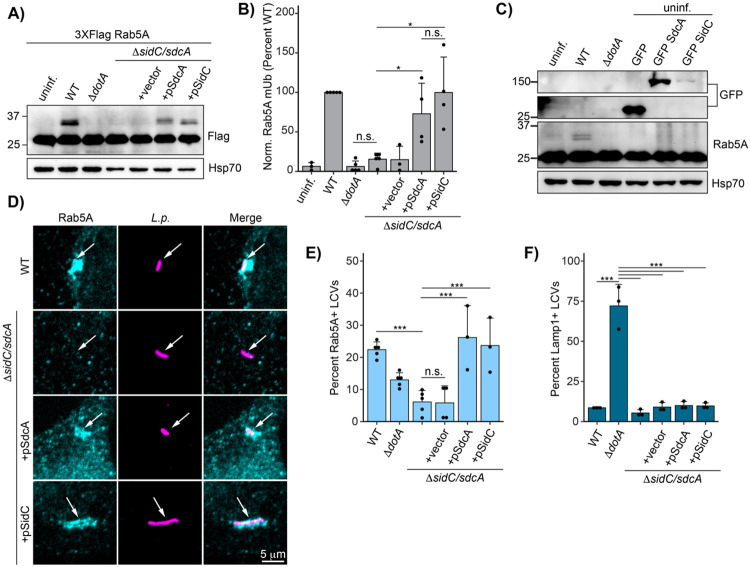
Fig 6: Bacterial effectors SidC/SdcA are required for Rab5A monoubiquitination and recruitment to the LCV.
(A) Immunoblot analysis of Rab5A monoubiquitination during infection with ΔsidC/sdcA L.p. strain panel (WT, ΔdotA, ΔsidC/sdcA, and ΔsidC/sdcA transformed with vector or plasmid expressing SdcA or SidC). HEK293T FcγR cells transfected with 3XFlag Rab5A were infected with the indicated strain or left uninfected. Cells were lysed at 4 hours post infection and probed with anti-Flag antibody. (B) Quantification of biological replicates (N=3-5) of experiment shown in (A). Normalized Rab5A monoubiquitination intensity was calculated as a percentage of WT L.p. infection levels (see Methods). Data was subjected to a one way ANOVA followed by a post-hoc Tukey-Kramer test for pairwise comparisons (* = p<0.05, n=3-4). (C) Immunoblot analysis of Rab5A monoubiquitination during SdcA or SidC ectopic expression. HEK293T FcγR were either left untransfected (lanes 1-3) or transfected with GFP alone or GFP-tagged SdcA or SidC for 24 hours. The untransfected cells were either left uninfected or infected with WT or ΔdotA L.p. for 4 hours. All cells were lysed and probed with anti-Flag and anti-GFP antibodies. (D) Representative images of Rab5A LCV recruitment levels for the ΔsidC/sdcA strain panel as observed by immunofluorescence. HeLa FcγR cells were infected with indicated strain for 1 hour, fixed, and probed with anti-Legionella and anti-Rab5A antibodies. (E) Quantification of biological replicates (N=3-5) of experiment shown in (D). 75-150 LCVs were scored per replicate as positive or negative for Rab5A recruitment, and the percent Rab5A+ LCVs was calculated per replicate. (F) Quantification of Lamp1 LCV recruitment for the ΔsidC/sdcA strain panel. HeLa FcγR cells were infected with indicated strain for 4 hr, fixed, and probed with anti-Legionella and anti-Lamp1 antibodies. LCVs were scored as in (E). For all graphs: bars represent mean value, error bars represent standard deviation. Individual points are values from each biological replicate. Statistical analysis of LCV scoring quantification: G test of independence was performed on pooled counts (positive vs. negative) from all biological replicates. Upon verifying significance (p<0.05), pairwise comparisons between strains were evaluated by post-hoc G-test using the Bonferroni-adjusted p-value as a significance threshold (p = 0.003). * = p<0.003, ** = p<0.0003, *** = p<0.00003, n.s. = p>0.003.