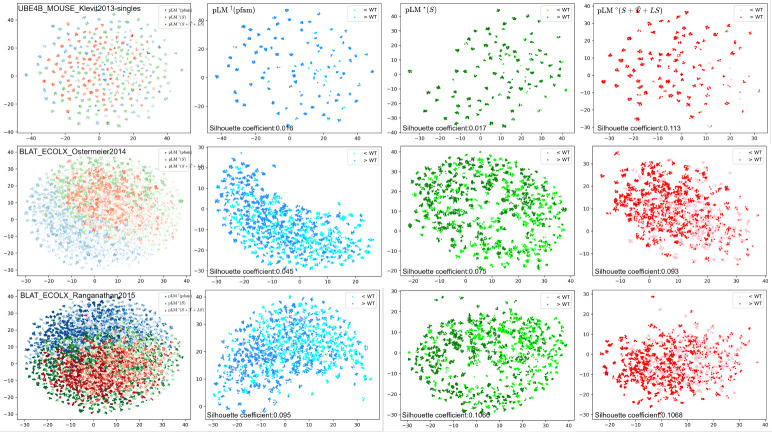
Fig. 5.
Low-dimension manifolds of variant embeddings by various pLMs (columns) for three representative proteins (rows) indicate that structure information led to better separation of high and low-fitness clusters. Whereas the first column is the union of three models: sequence pre-trained model (pLM†(pfam), blue), sequence fine-tuned model (pLM⋆(S), green), structure-informed model (SI-pLM⋄, red), each of the last three columns corresponds to one of the three models. In each figure, each point represents a variant, is located at the averaged embedding over mutant positions, and is colored according to the experimental fitness values (darker for higher fitness, continuous in the first column and binarized relative to the wild type in the other three columns).