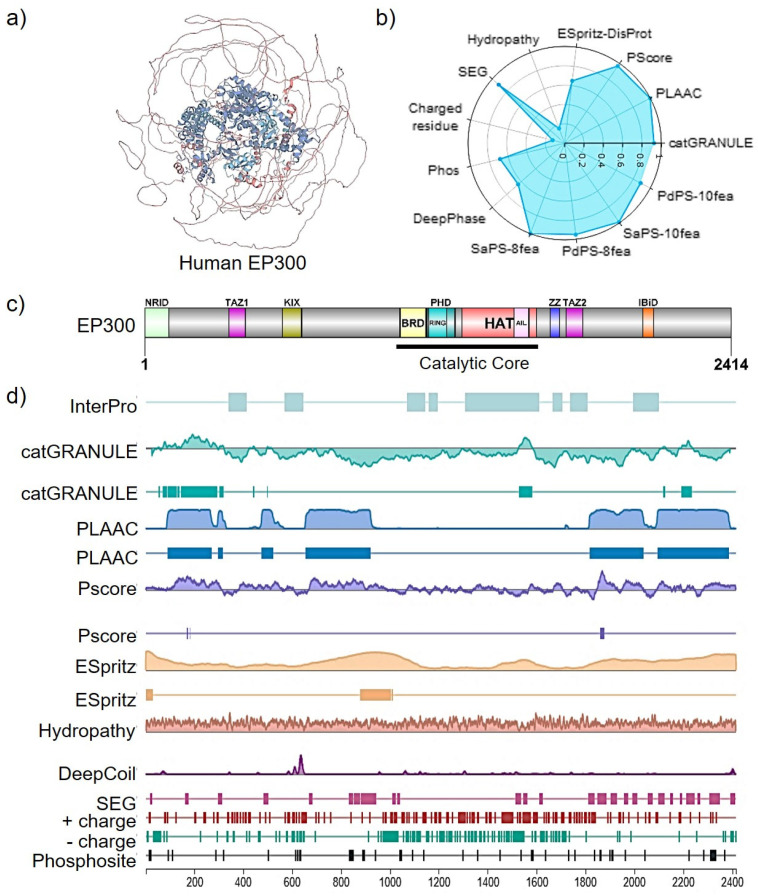
Figure 2.
EP300 structural insights (Uniprot Entry Q09472). (a) MobiDB manual annotation of intrinsic disorder domains (IDR, colored as alpha helices in blue) in EP300 3D folding derived from experimental data such as X-ray and NMR chemical shifts. (b) A radar chart displaying the ranking of disordered features for EP300 evaluated in the human proteome; ESpritz-DisProt: 0.648; Pscore: 0.968; PLAAC: 0.999; catGRANULE: 0.925; PsPS-10fea: 0.888; SaPS: 0.991; PdPS-8fea: 0.952; SaPS-8fea: 0.999; DeepPhase: 0.643; Phos: 0.688; Charged residue: 0.128; SEG: 0.911; Hydropathy: 0.162. A 1-ranking was shown for each feature value (the highest rank score is 1, and the lowest is 0). (c) Domain structure of human EP300. (d) PhaSePred protein feature viewer and LLPS-related predictions with residue-level scores. PLAAC (Prion-Like Amino Acid Composition); PS-Self score (proteins that can self-assemble to form condensates); PS-Part score (proteins whose phase separation behaviors are regulated by protein or nucleic acid partner components); SaPS (self-assembling phase-separating predictor); PdPS (partner-dependent phase-separating predictor); catGRANULE (granule-forming propensity predictor); PLD (prion-like domain); PScore (π-contact predictor); ESpritz (IDR predictor); SEG (low-complexity region, LCR); CIDER (hydropathy prediction); DeepCoil (coiled-coil domain predictor DeepCoil). Adapted from mobidb.org.