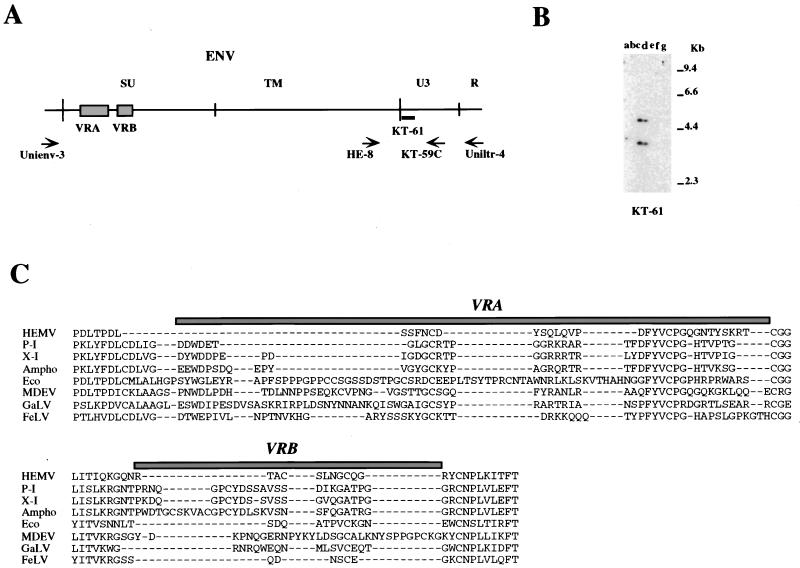
FIG. 6.
Detection of fragments of the M. spicelegus endogenous provirus (HEMV). (A) The locations of PCR primers and the oligonucleotide probes used. The approximate positions of the SU and TM regions of the env gene and the U3 and R regions of the LTR are indicated. Two hypervariable regions (VRA and VRB) in the SU region are also shown by boxes. (B) Detection of the HEMV provirus. Analysis of PvuII-digested mouse DNAs was performed by using the HEMV-specific oligonucleotide probe KT-61. Lanes: a, CZECH II/Ei (M. musculus); b, SPRET/Ei (M. spretus); c, PANCEVO/Ei (M. spicelegus); d, Halbturn (M. spicelegus); e, M. cervicolor; f, M. caroli; g, M. cookii. The approximate positions of molecular markers are also shown. (C) Amino acid sequences of the VRA and VRB regions of the HEMV env gene are aligned with the analogous regions of several type C retroviruses. Abbreviations and strains of the viruses included in this alignment are as follows: P-I (MX27) (49); X-I (NZB) (41); Ampho, amphotropic MLV (4070A) (42); Eco, ecotropic MLV (Akv) (58); MDEV, M. dunni endogenous virus (62); GalV, (U20589); FeLV, feline leukemia virus subgroup A (Glasgow-1) (48).