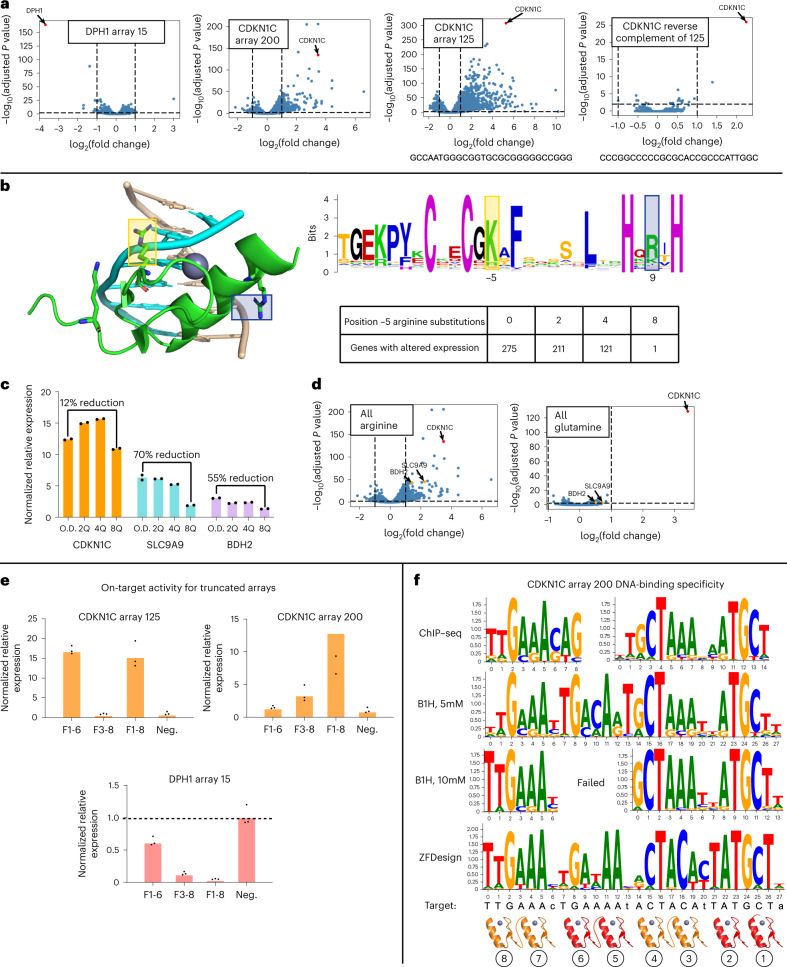
Fig. 6. ZF specificity and genome-wide activity.
a, Genome-wide RNA-seq results for CDKN1C arrays 125 and 200 and DPH1 array 15, and comparison with an array that binds the reverse complement of CDKN1C array 125. b, Left, structure of a ZF bound to DNA highlighting two potential phosphate contacts51. Right, the human ZF consensus with phosphate-contacting positions highlighted in yellow (−5) and blue (9)49. c, qPCR comparison for activation of the on-target CDKN1C gene as well as two off-target sequences with CDKN1C array 200 with between none and eight modifications at phosphate-contacting position −5. d, RNA-seq results for CDKN1C array 200 with arginines or glutamines at the −5 position of each ZF. e, On-target qPCR results for arrays with the N-terminal (F3–8) or C-terminal (F1–6) ZF pairs removed compared to an empty vector negative control (neg.). f, Specificity of CDKN1C array 200 array with glutamine at position −5 as determined by ChIP–seq, B1H selection at low (5 mM) and high (10 mM) stringency and specificity as predicted by ZFDesign. B1H specificity is a concatenation of the specificities determined for each of the two-finger pairs. ChIP–seq peaks contained two independent motifs, suggesting that the base-skipping linker allows modular, independent binding.