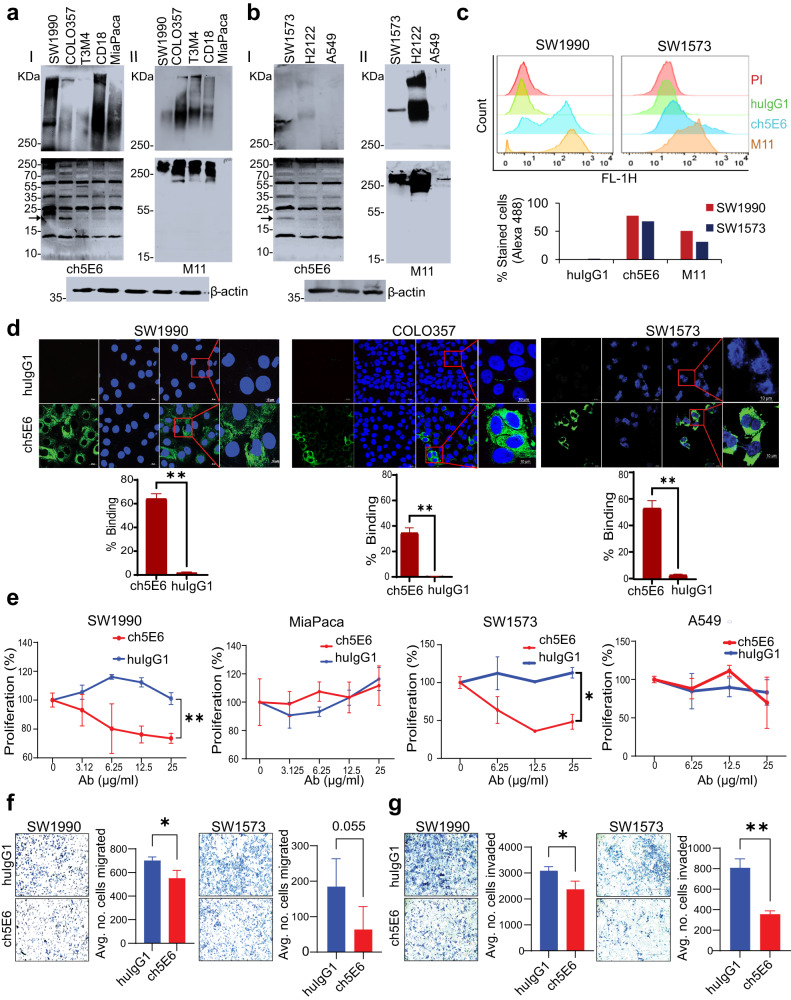
Fig. 2. ch5E6 recognizes endogenous forms of MUC16 in PC and NSCLC cell lines and inhibits their proliferation and metastasis.
Immunoblot analysis of ch5E6 binding to MUC16 in a PC (SW1990, COLO357, T3M4, CD18 and MIA PaCa-2) and b NSCLC cell line lysates (SW1573, H2122, and A549). MUC16 high molecular weight forms (HMW) were detected on 2%SDS-Agarose (I and II upper panel), and low molecular weight (LMW) forms on 12% SDS-PAGE electrophoresis (I and II lower panel). The same cell line lysates were also probed with an M11 (anti-CA125) antibody specific to the tandem repeat portion of MUC16. β-actin was run as a loading control. c Flow cytometry analysis of SW1990 and SW1573 cells for determining the surface binding potential of ch5E6. M11 and huIgG1 were used as positive and negative controls, respectively. d Confocal microscopy for analyzing specific binding of ch5E6 on MUC16 expressing SW1990, COLO357 (PC), and SW1573 (NSCLC) cell lines as compared to isotype control mAb huIgG1. Nuclei were stained with DAPI (Blue), and antibody binding was detected with fluorophore Alexa488 (green). % Staining on each cell line was calculated by measuring fluorescence (A488) intensity in 5 fields and normalization with DAPI. Scale bars, 100 µm; magnified images, 10 µm. e Anti-proliferative potential of ch5E6 in MUC16 expressing PC (SW1990, COLO357) and NSCLC (SW1573, H2122) cell lines. MUC16 negative line MIA PaCa-2 and A549 were used as a control in the experiment. The antibodies ch5E6 and huIgG1 were added at different concentrations from 0–25 μg/ml for 48 h. Real-time MT glo reagent (Promega) was used to detect the proliferation index. The data for COLO357 and H2122 is shown in Supplementary Fig. 2. The luminescence measurements were transformed to % proliferation. f inivasion and g migration of PC (SW1990) and NSCLC (SW1573) cells at 10 μg/ml were checked in Trans well insert assay with and without Matrigel coating, respectively. Cell numbers used are 1 × 106/ml in a 6-well trans well insert and 0.25 × 106/ml in a 24-well insert for invasion and migration assay, respectively. Experiments were performed in triplicates, and 10–20 images for each well were captured and counted. Error bars indicate SEM. *P < 0.05; **P < 0.01.