Abstract
Rhesus macaques immunized with simian immunodeficiency virus SIVmac239Δnef but not protected from SIVmac251 challenge were studied to determine the genetic and biological characteristics of the breakthrough viruses. Assessment of SIV genetic diversity (env V1-V2) revealed a reduction in the number of viral species in the immunized, unprotected macaques, compared to the number in nonimmunized controls. However, no evidence for selection of a specific V1-V2 genotype was observed, and biologically cloned isolates from the animals with breakthrough virus were similar with respect to replication kinetics and coreceptor use in vitro.
The ability of live, attenuated strains of simian immunodeficiency virus (SIV) to provide potent protection in macaques against a pathogenic SIV challenge has been documented by many groups (2, 3, 5, 6, 8, 14, 16, 18, 19, 23, 25–27). However, little is known about the genotype and phenotype of viruses that emerge in immunized macaques that fail to be protected, so-called “breakthrough viruses.” To gain insight into the mechanisms of protection, we studied cases where immunization with SIVmac239Δnef failed to protect against SIVmac251 challenge. Phenotypic analyses were also undertaken to determine patterns of coreceptor use and growth kinetics by using biologically cloned isolates obtained from the unprotected animals.
PCR analysis of nef sequences in immunized, unprotected animals.
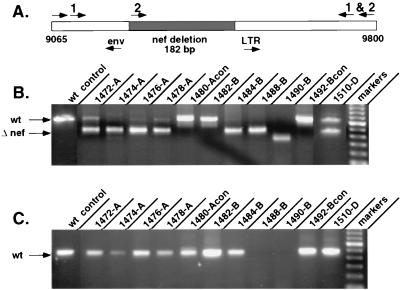
Sixteen female rhesus macaques were immunized with a live, attenuated SIV vaccine (SIVmac239Δnef) and challenged by the intravenous route with a pathogenic, uncloned stock of SIVmac251 either 5, 10, 15, or 25 weeks after immunization, as described earlier (6). To distinguish between the immunizing strain and the challenge virus, DNA PCR was performed by using specific primers that flank the deleted region of nef (Fig. 1) (6). By using one set of primers (Fig. 1A), sequences from both the wild-type (654-bp fragment) and Δnef (472-bp fragment) viruses were amplified. A second primer set (Fig. 1A) was used to increase detection of the wild-type nef product. One primer in this set hybridizes within the deleted region of SIVmac239Δnef, resulting in amplification of only wild-type nef sequences.
FIG. 1.
DNA PCR of the nef gene. (A) SIVmac239Δnef or SIVmac251 nef sequences were detected in PBMC samples obtained at 100 days postchallenge by using two sets of primer pairs (1 and 2). (B) Products amplified by primer set 1 showing both the Δnef fragment and the wild-type (wt) nef fragment. (C) Products amplified by primer set 2 showing only wild-type nef. Individual macaques are identified by the animal number and a letter representing the challenge group: A, 5-week challenge; B, 10-week challenge; D, 25-week challenge. 1480-A and 1492-B represent nonimmunized control animals in the 5- and 10-week challenge groups, respectively.
Of 16 immunized macaques, 7 were not protected from challenge based on evidence of an increase in plasma viremia, a decrease in CD4+ T cells, and the detection of SIVmac251 by PCR (6). To further characterize viruses present in the unprotected animals, DNA was extracted from peripheral blood mononuclear cell (PBMC) samples obtained 100 days after challenge and analyzed for the presence of either SIVmac239Δnef or SIVmac251 nef sequences. The results of these experiments demonstrate wild-type nef in four of four macaques challenged at 5 weeks (macaques 1472, 1474, 1476, and 1478 [Fig. 1C]), two of four challenged at 10 weeks (macaques 1482 and 1484 [Fig. 1C]), and one of four challenged at 25 weeks (macaque 1510 [Fig. 1C]) (6). In addition, a smaller, 472-bp PCR product was observed in six of seven macaques with breakthrough virus (macaques 1472, 1474, 1476, 1478, 1484, and 1510 [Fig. 1B]), indicating the persistence of SIVmac239Δnef. As expected, PCR analysis of the 5- and 10-week control animals (macaques 1480 and 1492 [Fig. 1B and C, respectively]) revealed only wild-type nef, consistent with SIVmac251 infection.
Genotypic analysis of the env V1-V2 region.
The V1-V2 region of SIV env was used to assess the genetic diversity of the breakthrough viruses. Preparation of viral RNA and DNA for genotypic analysis, as well as nested PCR amplification of the V1-V2 region, was performed as previously described (22). Three independently derived PCR products were pooled for analysis of viral quasispecies, thereby increasing the number of viral species within the PCR and reducing the possibility of sampling errors. Viral load in plasma at the time of heteroduplex mobility assay (HMA) sampling ranged from 1.5 × 104 to 5.4 × 105 RNA copies/ml on day 28 in the unprotected animals and from 8.6 × 104 to 1.6 × 106 RNA copies/ml on day 170, when lymph nodes were biopsied. In all cases, day 28 represented the earliest time at which SIVmac251 was detected in PBMC after challenge and plasma SIV RNA increased.
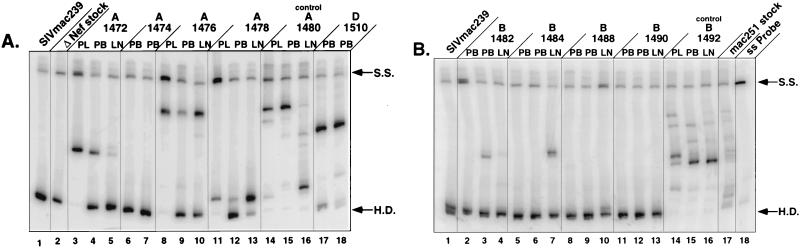
The amplified products were analyzed by a modified version of the HMA (9–11, 22). A single-stranded radiolabeled probe derived from the V1-V2 region of SIVmac239 was annealed to pooled, nonradiolabeled V1-V2 PCR product from each of the breakthrough viruses, from the SIVmac239Δnef stock used for immunization, and from the SIVmac251 challenge stock (Fig. 2). When the radiolabeled V1-V2 probe from SIVmac239 was hybridized with nonradiolabeled PCR product from SIVmac239, a single band was seen on polyacrylamide gels, representing a DNA homoduplex (Fig. 2A, lane 1). Similarly, a homoduplex was observed when the radiolabeled probe was annealed with V1-V2 PCR product from SIVmac239Δnef (Fig. 2A, lane 2), verifying that the virus used for immunization contained the same V1-V2 region as SIVmac239.
FIG. 2.
Single-stranded HMA of the env V1-V2 region. Genetic diversity in env V1-V2 was assessed by HMA with DNA extracted from infected PBMC (PB) (28 days postchallenge depicted for all; 100 days postchallenge depicted for macaques 1474, 1482, 1484, 1488, 1490, and 1510) or from lymph node (LN) (170 days postchallenge) or with plasma viral RNA (PL) (28 days postchallenge). (A) Animals 1472, 1474, 1476, and 1478 (5-week challenge group), animal 1480 (nonimmunized control), and animal 1510 (25-week challenge group); (B) macaques 1482, 1484, 1488, and 1490 (10-week challenge group) and macaque 1492 (nonimmunized control). S.S., single-stranded probe; H.D., homoduplex.
Genotypes that differ from the SIVmac239 probe form heteroduplexes that migrate with reduced mobility in polyacrylamide gels. Many distinct genotypes were observed with SIVmac251, representing viral variants present in the uncloned challenge stock (Fig. 2B, lane 17). Similarly, control macaques infected with uncloned SIVmac251, without prior immunization, had several replicating genotypes (macaques 1480 and 1492 [Fig. 2]). Although samples from 1480 showed fewer viral species replicating in PBMC and plasma than samples from 1492, multiple viral genotypes were observed in the lymph nodes (Fig. 2A), suggesting the presence of several replicating variants. This is consistent with previous results in experiments with three rhesus macaques inoculated with uncloned SIV (22). In each case, macaques infected by the intravenous route had a heterogenous quasispecies population in either lymph nodes, plasma, or PBMC, similar to the infecting virus stock (22).
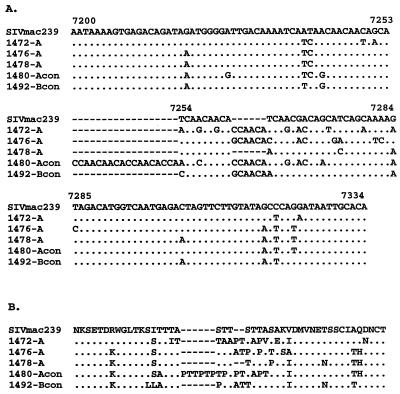
HMA analysis of the immunized animals included assessment of the V1-V2 genotypes found in plasma, PBMC, and lymph nodes (Fig. 2). The results of these experiments demonstrate a single band corresponding to the V1-V2 region of SIVmac239 in two macaques (1488 and 1490), indicating persistent infection with SIVmac239Δnef (Fig. 2B). The absence of additional bands strongly suggests that these animals were protected from infection with SIVmac251. These results are supported by the absence of wild-type nef sequences, as determined by nested DNA PCR (Fig. 1C). In contrast, breakthrough virus was detected in samples obtained from six other macaques (1472, 1476, 1478, 1482, 1484, and 1510). Each of these animals was also positive for wild-type nef by nested PCR (Fig. 1C). One other animal with breakthrough virus, 1474, was positive for wild-type nef by nested PCR (Fig. 1C), but HMA analysis of plasma and PBMC revealed only SIVmac239Δnef V1-V2 (Fig. 2A). Interestingly, only one or two genotypes were detected by HMA in the immunized, unprotected macaques, in contrast to the greater number of variants seen in nonimmunized controls (Fig. 2). This suggests that prior infection with SIVmac239Δnef can restrict the number of replicating genotypes in immunized animals in the absence of complete protection. However, the heteroduplex mobilities of the breakthrough viruses were different for each macaque, arguing against selection for a common V1-V2 genotype. Direct sequencing of the V1 region from three of the unprotected macaques (1472, 1476, and 1478) revealed no common V1 sequence (Fig. 3). Additional sequence data obtained from V2 (data not shown) and from biologically cloned viruses from two nonimmunized control animals (1480 and 1492) confirmed these observations (Fig. 3).
FIG. 3.
Sequence analysis of env V1 from macaques with breakthrough virus and control macaques. (A) Nucleotide sequences derived from PCR amplification and direct sequencing of the V1 region of env from immunized, unprotected macaques (1472, 1476, and 1478) and nonimmunized controls (animals 1480 and 1492); (B) predicted V1 amino acid sequences aligned to that of SIVmac239.
Biological cloning, coreceptor use, and replication of breakthrough viruses.
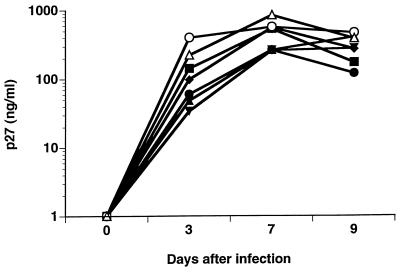
Biologically cloned isolates from the unprotected macaques were obtained by limiting-dilution PBMC cocultures, as previously described (7). Virus isolates were collected from the culture supernatants, while DNA was extracted from the infected cell pellets. The V1-V2 region was amplified by DNA PCR and analyzed by HMA. Individual clones were selected in which the viral genotype of the biological clone matched that of the breakthrough virus from the same animal when analyzed on HMA gels (data not shown). Biological clones representing the breakthrough viruses were obtained from four immunized, unprotected animals (1472, 1476, 1478, and 1510) and from two nonimmunized control animals (1480 and 1492). The isolates were evaluated to determine coreceptor use by inoculation into GHOST.4 cells expressing human CD4 and either rhesus CCR5 (4), rhesus CXCR4 (4), human GPR15 (BOB [12, 13, 15]), or human STRL33 (Bonzo [12]). GHOST.4 cells contain the gene for green fluorescent protein (GFP) under the control of the human immunodeficiency virus type 2 long terminal repeat (kindly provided by V. N. KewalRamani, New York University Medical Center), and infection was assessed by measuring the levels of GFP expression on day 3 after inoculation. The results of these experiments demonstrate that virus isolates from each of the immunized, unprotected and the nonimmunized control macaques use CCR5 for infection (Table 1). In addition, each of the biological clones used GPR15 and to a lesser extent STRL33, but none was able to infect cells expressing CXCR4; this is consistent with what has been reported for SIVmac239 (4, 12, 21) and SIVmac251 (4, 21). No differences in the pattern of coreceptor use among isolates from different animals were observed. In further experiments, each of the isolates was used to infect activated rhesus PBMC and virus replication was assessed by measuring SIV p27 in culture supernatants on days 3, 7, and 9 (Fig. 4). The results show that the isolates replicated to similar levels with approximately a twofold difference in the peak values between viruses isolated from immunized, unprotected macaques and from nonimmunized controls (366 ± 158 and 707 ± 221 ng/ml, respectively).
TABLE 1.
Coreceptor use by breakthrough viruses from immunized but unprotected macaques
| Animal no. | Level of GFPa for:
|
|||
|---|---|---|---|---|
| CCR5 | CXCR4 | GPR15 | STRL33 | |
| 1472 | +++ | − | ++ | + |
| 1476 | +++ | − | ++ | + |
| 1478 | +++ | − | ++ | + |
| 1482 | +++ | − | ++ | + |
| 1510 | +++ | − | ++ | + |
| 1480b | +++ | − | ++ | + |
| 1492b | +++ | − | ++ | + |
Coreceptor use was determined by measuring the levels of Tat-activated GFP in GHOST.4 cells expressing each coreceptor separately. +++, ++, +, and −, strong, moderate, weak, and negative fluorescence, respectively.
Nonimmunized control macaque infected with the SIVmac251 challenge virus.
FIG. 4.
Replication of biologically cloned viruses. Isolates from macaques with breakthrough virus (1472 [■], 1476 [●], 1478 [▴], 1482 [⧫], and 1510 [▾]) and control macaques (1480 [○] and 1492 [▵]) were obtained by limiting-dilution coculture (7) and used to infect activated rhesus PBMC in vitro. Virus replication was determined by measuring the levels of SIV p27 antigen on designated days after infection.
Discussion.
Earlier studies examined the temporal development of protective responses in 16 rhesus macaques immunized with SIVmac239Δnef (6). The results indicated that protection develops between 10 and 15 weeks after immunization and that this protection occurs in the absence of antibodies capable of neutralizing the primary challenge virus in vitro (6). Of 16 rhesus macaques immunized with SIVmac239Δnef, 7 were not protected from SIVmac251 challenge. All but one of these macaques were challenged after the peak of viremia (5 to 10 weeks), when plasma virus was low and seroconversion had occurred (6). In the present study, we examined the genotype and phenotype of viruses replicating in the unprotected macaques. Because these animals were challenged with an uncloned stock of SIVmac251, we were able to assess genetic diversity within the V1-V2 region of env using a modified HMA (9–11, 22). This region has been shown to be one of the more variable regions of the SIV genome (1, 17, 20, 24). The number of viral genotypes, as determined by HMA analysis, indicated that fewer viral species from the challenge stock were replicating within the immunized, unprotected macaques than in the nonimmunized controls, suggesting that some selection had occurred as a result of immunization. To address this in part, we evaluated genetic diversity in other regions of the SIV genome (env V3 to V5, gag, nef) by HMA and found no diversity within these additional sites (data not shown). Alternatively, it is possible that differences in viral diversity may have resulted from different kinetics of viral evolution following infection of immunized and control animals. However, we observed a significant reduction in viral diversity by day 28 (the earliest day tested after challenge) in the immunized, unprotected macaques, and further sampling of PBMC (day 100) and lymph nodes (day 170) did not reveal any significant changes in the patterns of diversity. Therefore, we favor the hypothesis that a restriction in viral diversity occurred early after challenge in unprotected macaques and may reflect preexisting antiviral immunity induced by immunization with the live, attenuated SIV vaccine. Based on HMA analysis and sequencing data, there was no evidence for selection of a specific V1-V2 viral genotype among the animals with breakthrough virus, and we found no evidence to suggest that the viruses replicating in these animals were distinct with respect to coreceptor use or growth kinetics in vitro. Therefore, no common genotypic or phenotypic property could be attributed to the breakthrough viruses among the parameters studied here. Further experiments will be needed to identify factors that are involved in reducing the number of replicating variants in the immunized, unprotected animals and allowing penetration and replication of the breakthrough strains.
Nucleotide sequence accession numbers.
The sequences of 1472-A, 1476-A, 1478-A, 1480-Acon, and 1492-Bcon have been deposited in GenBank under accession no. AF129452, AF129453, AF129454, AF129455, and AF129456, respectively.
Acknowledgments
We thank Agegnehu Gettie for providing blood samples and Eric Delwart for helpful discussions.
This work was supported by the NIH (AI28147 and AI36598), the Aaron Diamond Foundation, and an Aaron Diamond Foundation Fellowship awarded to D.L.S.
REFERENCES
- 1.Almond N, Jenkins A, Heath A B, Kitchin P. Sequence variation in the env gene of simian immunodeficiency virus recovered from immunized macaques is predominantly in the V1 region. J Gen Virol. 1993;74:865–871. doi: 10.1099/0022-1317-74-5-865. [DOI] [PubMed] [Google Scholar]
- 2.Almond N, Kent K, Cranage M, Rud E, Clarke B, Stott E J. Protection by attenuated simian immunodeficiency virus in macaques against challenge with virus-infected cells. Lancet. 1995;345:1342–1344. doi: 10.1016/s0140-6736(95)92540-6. [DOI] [PubMed] [Google Scholar]
- 3.Beer B, Baier M, Megede J Z, Norley S, Kurth R. Vaccine effect using a live attenuated nef-deficient simian immunodeficiency virus of African green monkeys in the absence of detectable vaccine virus replication in vivo. Proc Natl Acad Sci USA. 1997;94:4062–4067. doi: 10.1073/pnas.94.8.4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Z, Zhou P, Ho D D, Landau N R, Marx P A. Genetically divergent strains of simian immunodeficiency virus use CCR5 as a coreceptor for entry. J Virol. 1997;71:2705–2714. doi: 10.1128/jvi.71.4.2705-2714.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clements J E, Montelaro R C, Zink M C, Amedee A M, Miller S, Trichel A M, Jagerski B, Hauer D, Martin L N, Bohm R P, Murphey-Corb M. Cross-protective immune responses induced in rhesus macaques by immunization with attenuated macrophage-tropic simian immunodeficiency virus. J Virol. 1995;69:2737–2744. doi: 10.1128/jvi.69.5.2737-2744.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Connor R I, Montefiori D C, Binley J M, Moore J P, Bonhoeffer S, Gettie A, Fenamore E A, Sheridan K E, Ho D D, Dailey P J, Marx P A. Temporal analyses of virus replication, immune responses, and efficacy in rhesus macaques immunized with a live, attenuated simian immunodeficiency virus vaccine. J Virol. 1998;72:7501–7509. doi: 10.1128/jvi.72.9.7501-7509.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Connor R I, Notermans D W, Mohri H, Cao Y, Ho D D. Biological cloning of functionally diverse quasispecies of HIV-1. AIDS Res Hum Retroviruses. 1993;9:541–546. doi: 10.1089/aid.1993.9.541. [DOI] [PubMed] [Google Scholar]
- 8.Daniel M D, Kirchhoff F, Czajak S C, Sehgal P K, Desrosiers R C. Protective effects of a live attenuated SIV vaccine with a deletion in the nef gene. Science. 1992;258:1938–1941. doi: 10.1126/science.1470917. [DOI] [PubMed] [Google Scholar]
- 9.Delwart E, Shpaer E G, Louwagie J, McCutchan F E, Grez M, Rubsamen-Waigmann H, Mullins J I. Genetic relationships determined by a DNA heteroduplex mobility assay: analysis of HIV-1 env genes. Science. 1993;262:1257–1261. doi: 10.1126/science.8235655. [DOI] [PubMed] [Google Scholar]
- 10.Delwart E L, Pan H, Sheppard H W, Wolpert D, Neumann A U, Korber B, Mullins J I. Slower evolution of human immunodeficiency virus type 1 quasispecies during progression to AIDS. J Virol. 1997;71:7498–7508. doi: 10.1128/jvi.71.10.7498-7508.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delwart E L, Sheppard H W, Walker B D, Goudsmit J, Mullins J I. Human immunodeficiency virus type 1 evolution in vivo tracked by DNA heteroduplex mobility assays. J Virol. 1994;68:6672–6683. doi: 10.1128/jvi.68.10.6672-6683.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng H K, Unutmaz D, KewelRamani V N, Littman D R. Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature. 1997;388:296–300. doi: 10.1038/40894. [DOI] [PubMed] [Google Scholar]
- 13.Liao F, Alkahtib G, Peden K W C, Sharma G, Berger E A, Farber J M. STRL33, a novel chemokine receptor-like protein, functions as a fusion cofactor for both macrophage-tropic and T cell line-tropic HIV-1. J Exp Med. 1997;185:2015–2023. doi: 10.1084/jem.185.11.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lohman B L, McChesney M B, Miller C J, McGowan E, Joye S M, Van Rompay K K A, Reay E, Antipa L, Pedersen N C, Marthas M L. A partially attenuated simian immunodeficiency virus induces host immunity that correlates with resistance to pathogenic virus challenge. J Virol. 1994;68:7021–7029. doi: 10.1128/jvi.68.11.7021-7029.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marcon L, Choe H, Martin K A, Farzan M, Ponath P D, Wu L, Newman W, Gerard N, Gerard C, Sodroski J. Utilization of C-C chemokine receptor 5 by the envelope glycoproteins of a pathogenic simian immunodeficiency virus, SIVmac239. J Virol. 1997;71:2522–2527. doi: 10.1128/jvi.71.3.2522-2527.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marthas M L, Sutjipto S, Miller C J, Higgins J, Torten J, Unger R E, Marx P A, Pedersen N C. Efficacy of live-attenuated and whole-inactivated SIV vaccines against intravenous and vaginal challenge. Vaccines. 1992;92:117–121. [PubMed] [Google Scholar]
- 17.Neildez O, Le Grand R, Caufour P, Vaslin B, Chéret A, Matheux F, Théodoro F, Roques P, Dormont D. Selective quasispecies transmission after systemic or mucosal exposure of macaques to simian immunodeficiency virus. Virology. 1998;243:12–20. doi: 10.1006/viro.1997.9026. [DOI] [PubMed] [Google Scholar]
- 18.Norley S, Beer B, Binninger-Schinzel D, Cosma C, Kurth R. Protection from pathogenic SIVmac challenge following short-term infection with a nef-deficient attenuated virus. Virology. 1996;219:195–205. doi: 10.1006/viro.1996.0237. [DOI] [PubMed] [Google Scholar]
- 19.Otsyula M G, Miller C J, Tarantal A F, Marthas M L, Greene T P, Collins J R, Van Rompay K K A, McChesney M B. Fetal or neonatal infection with attenuated simian immunodeficiency virus results in protective immunity against oral challenge against pathogenic SIVmac251. Virology. 1996;222:275–278. doi: 10.1006/viro.1996.0420. [DOI] [PubMed] [Google Scholar]
- 20.Overbaugh J, Rudensey L M, Papenhausen M D, Benveniste R E, Morton W R. Variation in simian immunodeficiency virus env is confined to V1 and V4 during progression to simian AIDS. J Virol. 1991;65:7025–7031. doi: 10.1128/jvi.65.12.7025-7031.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rucker J, Edinger A L, Sharron M, Samson M, Lee B, Berson J F, Yi Y, Margulies B, Collman R G, Doranz B J, Parmentier M, Doms R W. Utilization of chemokine receptors, orphan receptors, and herpesvirus-encoded receptors by diverse human and simian immunodeficiency viruses. J Virol. 1997;71:8999–9007. doi: 10.1128/jvi.71.12.8999-9007.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sodora D L, Lee F, Dailey P J, Marx P A. A genetic and viral load analysis of the simian immunodeficiency virus during the acute phase in macaques inoculated by the vaginal route. AIDS Res Hum Retroviruses. 1998;14:171–181. doi: 10.1089/aid.1998.14.171. [DOI] [PubMed] [Google Scholar]
- 23.Stahl-Hennig C, Dittmer U, NiBlein T, Petry H, Jurkiewicz E, Fuchs D, Wachter H, Matz-Rensing K, Kuhn E M, Kaup F J, Rud E W, Hunsmann G. Rapid development of vaccine protection in macaques by live-attenuated simian immunodeficiency virus. J Gen Virol. 1996;77:2969–2981. doi: 10.1099/0022-1317-77-12-2969. [DOI] [PubMed] [Google Scholar]
- 24.Tao B, Fultz P N. Molecular and biological analyses of quasispecies during evolution of a virulent simian immunodeficiency virus, SIVsmmPBj14. J Virol. 1995;69:2031–2037. doi: 10.1128/jvi.69.4.2031-2037.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Titti F, Sernicola L, Geraci A, Panzini G, DiFabio S, Belli R, Monardo F, Borsetti A, Maggiorella M T, Koanga-Mogtomo M, Corrias F, Zamarchi R, Amadori A, Chieco-Bianchi L, Verani P. Live attenuated simian immunodeficiency virus prevents super-infection by cloned SIVmac251 in cynomolgus monkeys. J Gen Virol. 1997;78:2529–2539. doi: 10.1099/0022-1317-78-10-2529. [DOI] [PubMed] [Google Scholar]
- 26.Wyand M S, Manson K H, Garcia-Moll M, Montefiori D, Desrosiers R C. Vaccine protection by a triple deletion mutant of simian immunodeficiency virus. J Virol. 1996;70:3724–3733. doi: 10.1128/jvi.70.6.3724-3733.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wyand M S, Manson K H, Lackner A A, Desrosiers R C. Resistance of neonatal monkeys to live attenuated vaccine strains of simian immunodeficiency virus. Nat Med. 1997;3:32–36. doi: 10.1038/nm0197-32. [DOI] [PubMed] [Google Scholar]