Figure 4. Association of 3D rewiring with cell-type specific gene expression.
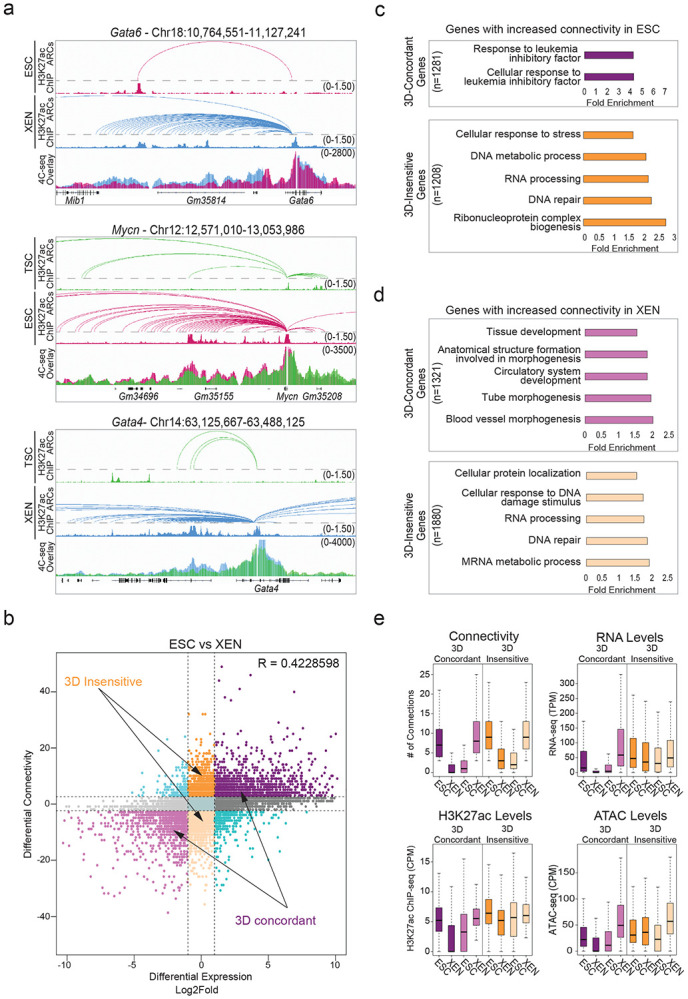
a. Examples of pairwise comparisons documenting 3D rewiring at developmental genes in TSC, ESC and XEN as detected by H3K27ac HiChIP (shown as arcs on top along with the respective H3K27ac ChIP-seq tracks) and validated by independent 4C-seq experiments (merged tracks at the bottom). Averaged 4C-seq signals from three biological replicates are presented after normalization to the sequencing depth.
b. Correlation between differential HiChIP connectivity/hubness and differential gene expression in ESC vs XEN cells. R represents Spearman correlation identifies distinct groups of genes. We focus on the two most prominent groups: 3D-insensitive genes, defined as genes with differential connectivity >3 but no transcriptional changes (log2FC<1 or >−1) and 3D-concordant genes for which connectivity and expression changes (log2FC >1 or <−1) positively correlate (Supplementary Table 5).
c. Gene ontology analysis depicting the most significant biological processes enriched in the 3D concordant (purple) and 3D insensitive (orange) groups in ESC cells as defined in (b). All genes in A compartments were used as background. For further details see also Supplementary Table 3.
d. Same as in (c), but for genes with increased connectivity in XEN. For further details see also Supplementary Table 3.
e. Comparison of connectivity, gene expression levels (TPM) as well as H3K27ac and ATAC CPM levels on the promoters of 3D-concordant and 3D-insensitive genes in ESC and XEN cells. Insensitive genes show higher levels of connectivity, H3K27ac, ATAC and expression in both cell types. Wilcoxon rank sum test was used for all comparisons (Supplementary Table 9).
Note: all statistics are provided in Supplementary Table 9.
