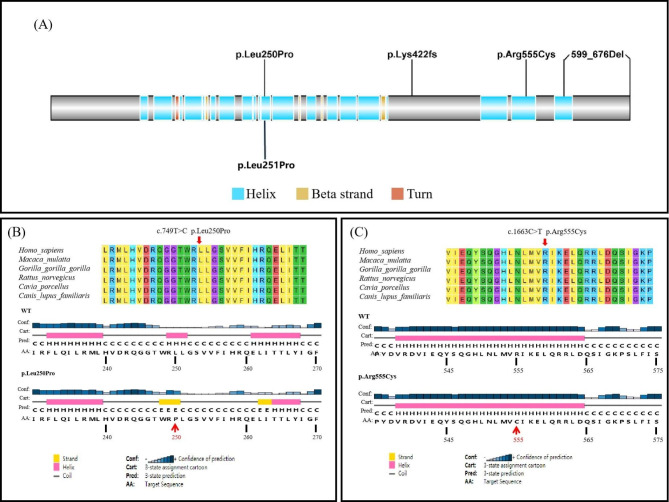
Fig. 2.
(A) The illustration indicated the amino acid changes in KCNQ1 found in our study. (B) KCNQ1 p.Leu250Pro was located in a highly conserved region across all available species evolutionally (upper). Altered secondary structure of p.Leu250Pro mutant protein was predicted when compared with the wild type (the altered residue was shown with red arrow) (lower). (C) KCNQ1 p.Arg555Cys was also located in a highly conserved region across all available species evolutionally (upper). No significant alteration of the secondary structure of p.Arg555Cys mutant protein was predicted when compared with the wild type (lower, the altered residue is shown with red arrow) (lower)