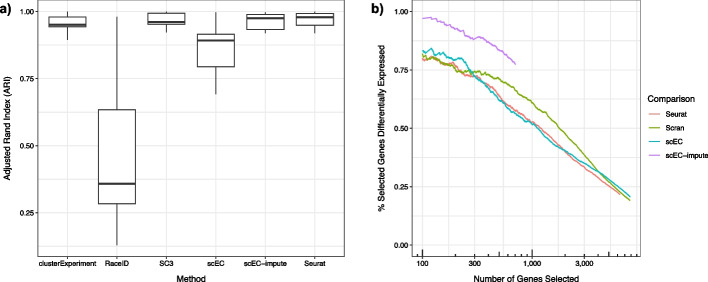
Fig. 5.
Benchmarking of scEC performance in unsupervised clustering and feature selection. a Adjusted Rand Index of clusterings produced by specified methods against known ground truth for seven data sets, each consisting of three or five cancerous lines sequenced on different platforms. With an additional imputation step, scEC performs on par with other methods. b The percent of the top N genes by different feature selection metrics that are differentially expressed. Data set is Sc-seq from three cancerous cell lines sequenced by Drop-seq (with 2005 differential expressed genes identified from non-parametric testing for each cell line versus the remaining; Wilcox test, fasle discovery rate corrected p-value ). The greater ability of scEC-impute to a priori select differentially expressed genes is repeated across each benchmark data set, see Additional file 1: Fig. S2. Note that the imputation step in scEC-impute assigns many genes a heterogeneity I(g) of zero, resulting in a low cut-off on total number of selectable genes