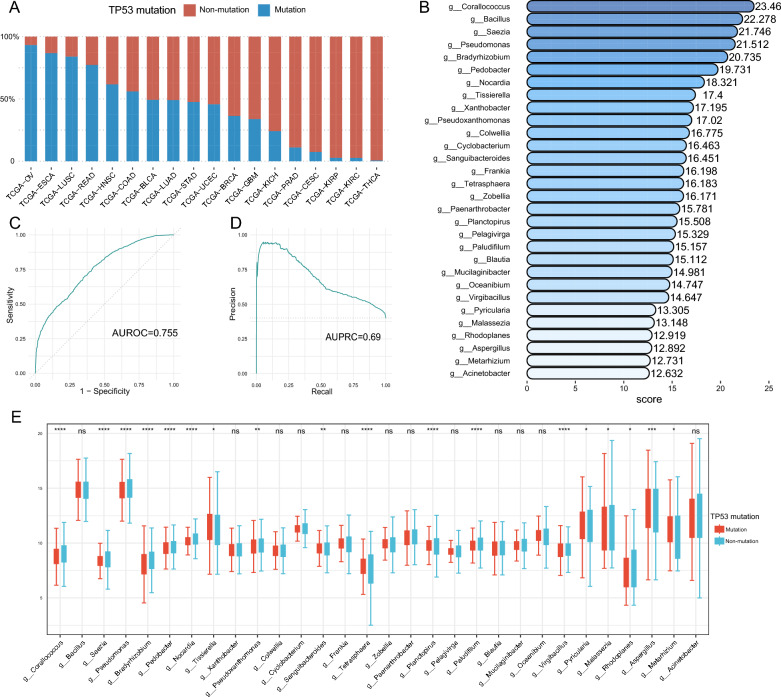
Fig. 6.
Tumor microbiome can distinguish TP53 mutations. A The proportion (y-axis) of tumors with TP53 mutations (blue) and TP53 non-mutations (red) in OV, ESCA, LUSC, READ, HNSC, COAD, BLCA, LUAD, STAD, UCEC, BRCA, GBM, KICH, PRAD, CESC, KIRP, KIRC and THCA. B Top 30 tumor microbiomes with highest ML model importance score (x-axis). The darker the color, the higher the importance score. C AUROC value of the ML model. D AUPRC value of the ML model. E Distribution of the top 30 tumor microbiome of highest ML model importance in TP53 mutations (red) and TP53 non-mutations (blue). The y-axis indicates the relative abundance of microorganisms. Statistical tests were performed using the rank sum test. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001,ns P > 0.05. TCGA the cancer genome atlas, OV ovarian cancer, ESCA esophageal cancer, LUSC lung squamous cell carcinoma, READ rectal cancer, HNSC head and neck cancer, COAD colon cancer, BLCA bladder cancer, LUAD lung adenocarcinoma, STAD stomach cancer, UCEC endometrioid cancer, BRCA breast cancer, GBM glioblastoma, KICH kidney chromophobe, PRAD prostate cancer, CESC cervical cancer, KIRP kidney papillary cell carcinoma, KIRC kidney clear cell carcinoma, THCA thyroid cancer, TP53 Tumor Protein P53; ML machine learning, AUROC the area under the curve of the receiver operating characteristic, AUPRC the area under the curve of the precision-recall, ns no significance