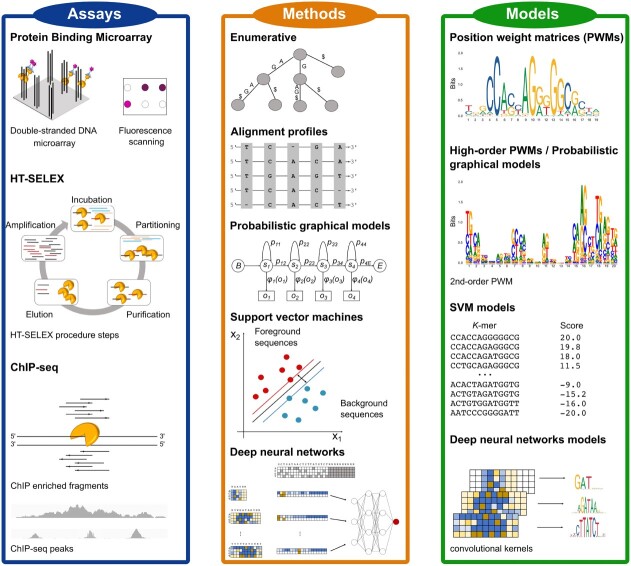
Figure 1.
Experimental and computational methods to discover TFBS and popular models to represent binding site motifs. Protein binding microarray (PBM), HT-SELEX and ChIP-seq have become the most popular assays to determine TF binding preferences and identify their target sites (TFBS) in recent years. Computational motif discovery methods can be grouped into five classes, based on the algorithms employed to discover TFBS: enumerative, alignment-based, probabilistic graphical model-based, SVM-based and DNN-based methods. TFBS sequences prioritized by motif discovery algorithms are encoded in computational models representing the binding preferences of the investigated TFs.