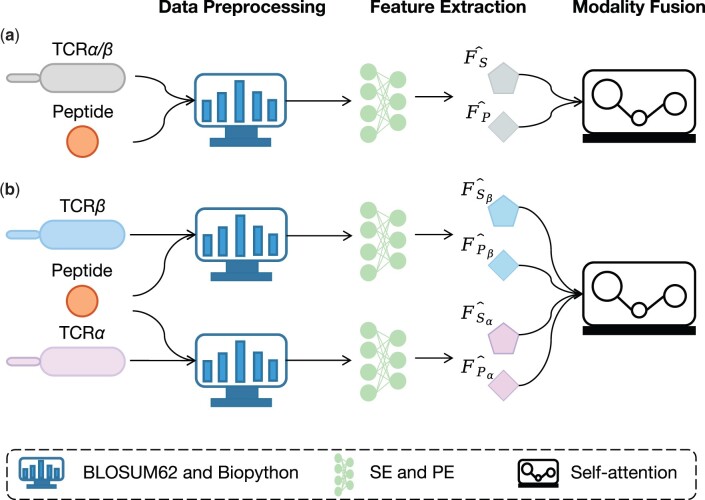
Figure 1.
The framework of MIX-TPI to handle both (a) single-chain TCR data, i.e. TCR (-chain) and (b) paired TCR data, i.e. both TCR and TCR. TCRs and peptide sequences are firstly fed into the data preprocessing module, where they are encoded with BLOSUM62 and Biopython (Cock et al. 2009) libraries, respectively, to generate the embedding matrices of SE and PE. Subsequently, SE and PE are used to extract sequence and physicochemical features, respectively, with CNNs. Finally, the modality-invariant and modality-specific representations are learned and passed to the self-attention fusion layer to predict the TCR–pMHC interactions.