Figure 1: Development and design of detection rules for gutSMASH.
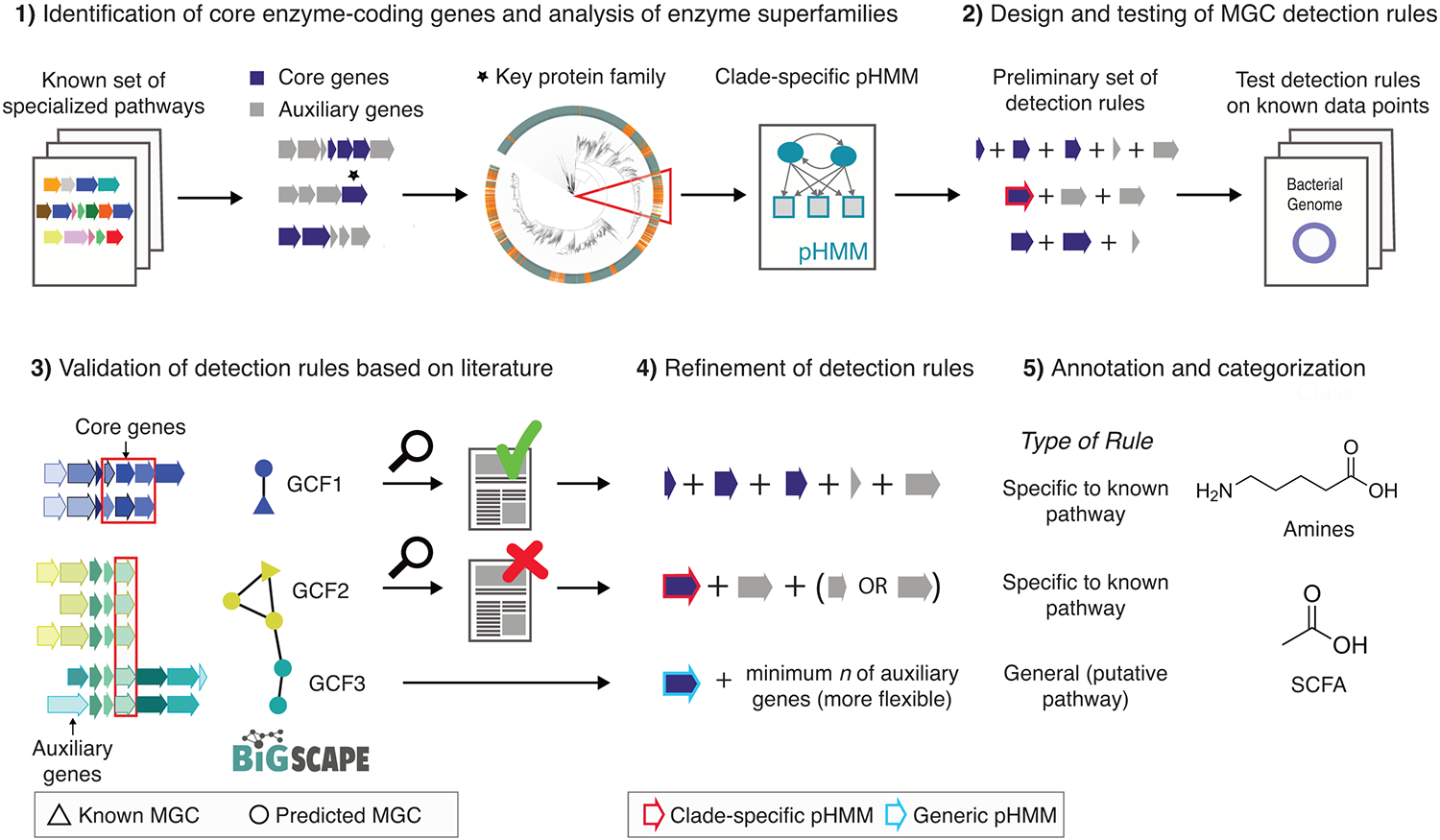
(1) A set of known and characterized MGC-encoded pathways were curated from the literature. Protein domains were identified across all MGCs and core enzymatic domains were manually identified. For enzymatic domains belonging to broad multifunctional enzyme families, protein superfamily phylogenies were built to create clade-specific pHMMs. (2) These domains were incorporated in the initial detection rules. The detection rules were run on a test set, and all the MGC predicted by the same rule were grouped together and (3) run through BiG-SCAPE, which grouped the MGCs into gene cluster families (GCFs). (4) Based on literature analysis of GCF members, detection rules were manually fine-tuned to either include or exclude MGC architectures that were either related to specialized primary metabolism or not. (5) Finally, fine-tuned detection rules were annotated and categorized into different MGC classes based on their metabolic end products.
