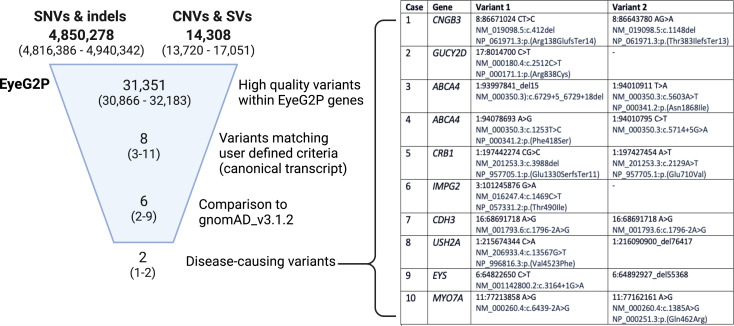
Figure 3.
EyeG2P effectively prioritises pathogenic variation from whole genome sequencing data sets. The median and range of genomic variants identified across 10 individuals with whole genome sequencing analysis are shown at each stage of the filtering process. The complete list of clinically reported pathogenic and likely pathogenic variants prioritised by EyeG2P as a cause of inherited ophthalmic disorders is shown (genomic coordinates refer to the GRCh38 genome build). gnomAD, Genome Aggregation Database; indels, small insertion and deletion events; SNVs, single nucleotide variant; SVs, structural variants.