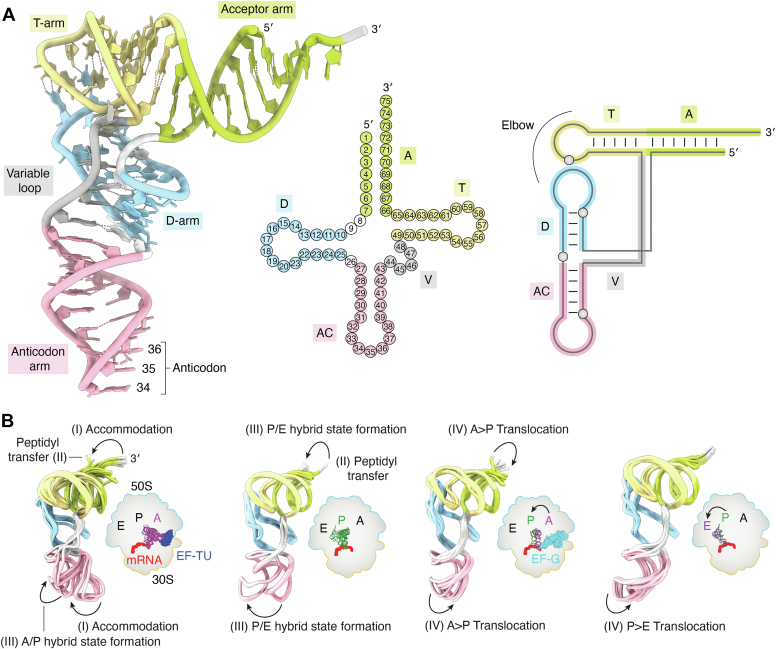
Figure 1.
The structure of tRNA at different levels and during translation.A, left: specific regions are indicated on yeast tRNAPheGAA (PDB ID 1EHZ) as follows: 3ʹ and 5ʹ end; acceptor arm (light green); T-arm (yellow); D-arm (light blue); variable loop (gray); anticodon arm (pink); and 34, 35, 36 anticodon nt. The dimensions of folded tRNAs are approximately 60 × 70 × 25 Å. Middle: clover-leaf secondary structure representation of tRNA numbered according to the standard convention. Right: tertiary nucleotide interactions responsible for the L-shape tRNA structure. B, multiple structures of tRNA extracted from the context of elongating bacterial 70S ribosome determined by time-resolved cryo-EM. The schematic demonstrates local flexibility of the A-site (magenta), P-site (dark green), and E-site (purple) tRNA associated with the cognate mRNA (red) bound to the 70S ribosome [large 50S (blue) and small 30S (orange)]. The arrows highlight the bending of the 3ʹ end (silver) from the acceptor arm (light green) and the swinging of the anticodon arm (pink). The major sequential steps of the elongation cycle associated with the L-shape distortion are indicated with Roman numerals (I–IV). The two superimpositions on the left (A-site phenylalanine- and P-site methionine-tRNA) follow the steps and changes after cognate aminoacyl-tRNA delivery by elongation factor EF-TU (dark blue). The two superimpositions on the right depict AP proline- and PE methionine-tRNA translocation facilitated by elongation factor EF-G (cyan), which resets the system for the next elongation cycle. EF-Tu, Elongator factor-Tu.