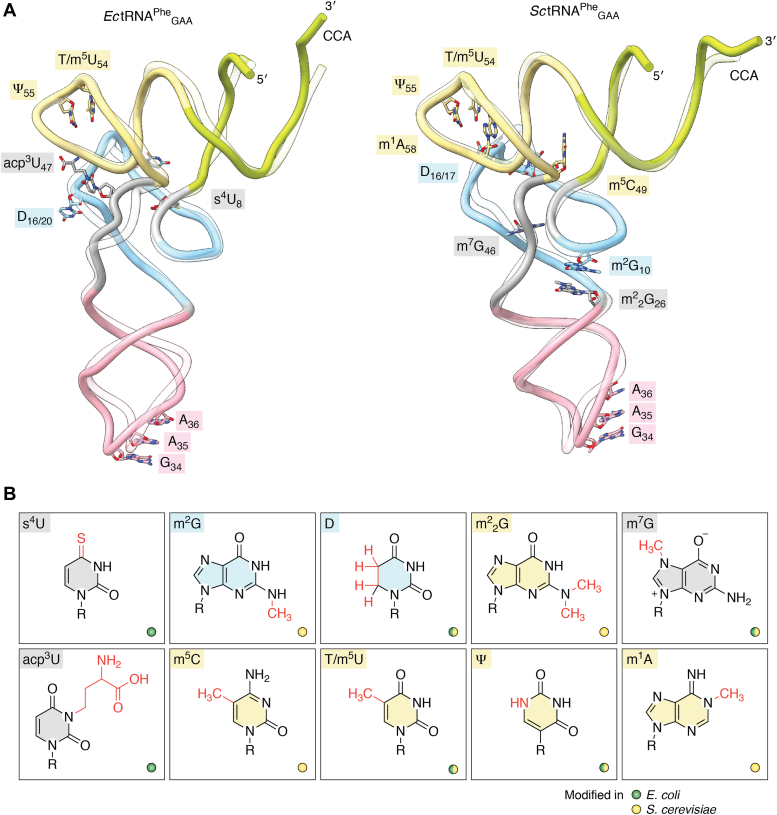
Figure 2.
Structure of unmodified and modified tRNAPhe.A, comparison of prokaryotic and eukaryotic tRNAPheGAA. Superimpositions of in vitro transcribed (transparent) and endogenous (opaque) tRNAPheGAA from Escherichia coli (Ec) and Saccharomyces cerevisiae (Sc) obtained by crystallography. These tRNAs are depicted as cartoons using the same color code for each region as in Figure 1. Modifications that are important for the tRNA core structure are represented in sticks and colored according to their position in the tRNA. The PDB IDs and resolution limits of each tRNA are as follows: unmodified EctRNA (PDB ID 3L0U at 3.0 Å) and SctRNA (PDB ID 5AXM at 2.2 Å) and modified EctRNA (PDB ID 6Y3G at 3.1 Å) and SctRNA (PDB ID 1EHZ at 1.9 Å). The m7G47 modification in EctRNA (PDB ID 6Y3G) is present in tRNAPheGAA but is not visible in the electron density map. B, chemical structures of modified nucleosides that are shown in (A). The modifications are indicated in red and the ribose moiety is annotated with R. Base colors correspond to the modified target positions in (A), while the color (green or yellow) of the small circle indicates the presence of modification in E. coli or S. cerevisiae. RMSD values for E. coli tRNAPhe and S. cerevisiae tRNAPhe were calculated with ChimeraX using the backbone atoms of nt 3 to 74. E. coli: overall RMSD 2.669 Å2, acceptor stem 4.426 Å2, D-arm 1.595 Å2, anticodon-loop 2.716 Å2, variable loop 2.362 Å2, T-arm 1.349 Å2; S. cerevisiae: overall RMSD 1.662 Å2, acceptor stem 2.132 Å2, D-arm 2.194 Å2, anticodon-loop 1.628 Å2, variable loop 0.801 Å2, T-arm 0.665 Å2.