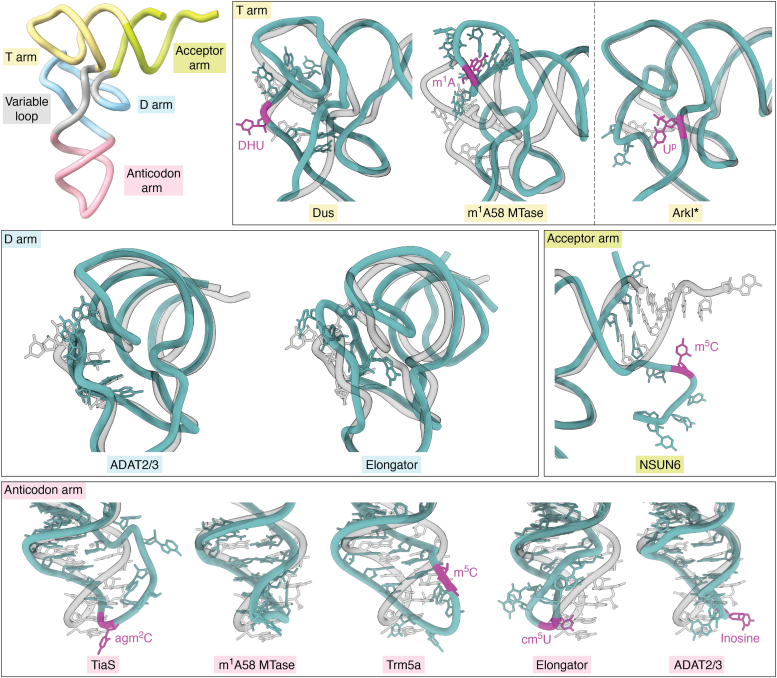
Figure 4.
Modification enzymes influence tRNA structure in various regions. Yeast tRNAPheGAA (PDB ID 1EHZ) is aligned on each structure by the matchmaker tool in ChimeraX and shown transparently. Structures are sorted by the region affected structurally, and the names of the modifying enzymes are colored, accordingly. Modification sites are marked in magenta. Structures of full length-tRNAs are shown without their respective protein partners - Dus (PDB ID 3B0V), m1A58 MTase (PDB ID 5CD1), Elongator (PDB ID 8ASW), ADAT2/3 (PDB ID 8AW3), TiaS (PDB ID 3AMT), Trm5a (PDB ID 5WT3), NSUN6 (PDB ID 5WWT). ArkI (PDB ID 7VNV) is a crystal structure of tRNA alone, modified by TkArkI.