Abstract
Identifying tumor DNA mismatch repair deficiency (dMMR) is important for precision medicine. Tumor features, individually and in combination, derived from whole-exome sequenced (WES) colorectal cancers (CRCs) and panel-sequenced CRCs, endometrial cancers (ECs), and sebaceous skin tumors (SSTs) were assessed for their accuracy in detecting dMMR. CRCs (n = 300) with WES, where mismatch repair status was determined by immunohistochemistry, were assessed for microsatellite instability (MSMuTect, MANTIS, MSIseq, and MSISensor), Catalogue of Somatic Mutations in Cancer tumor mutational signatures, and somatic mutation counts. A 10-fold cross-validation approach (100 repeats) evaluated the dMMR prediction accuracy for i) individual features, ii) Lasso statistical model, and iii) an additive feature combination approach. Panel-sequenced tumors (29 CRCs, 22 ECs, and 20 SSTs) were assessed for the top performing dMMR predicting features/models using these three approaches. For WES CRCs, 10 features provided >80% dMMR prediction accuracy, with MSMuTect, MSIseq, and MANTIS achieving ≥99% accuracy. The Lasso model achieved 98.3% accuracy. The additive feature approach, with three or more of six of MSMuTect, MANTIS, MSIseq, MSISensor, insertion-deletion count, or tumor mutational signature small insertion/deletion 2 + small insertion/deletion 7 achieved 99.7% accuracy. For the panel-sequenced tumors, the additive feature combination approach of three or more of six achieved accuracies of 100%, 95.5%, and 100% for CRCs, ECs, and SSTs, respectively. The microsatellite instability calling tools performed well in WES CRCs; however, an approach combining tumor features may improve dMMR prediction in both WES and panel-sequenced data across tissue types.
DNA mismatch repair (MMR) deficiency (dMMR) is an important molecular phenotype of solid tumors characterized by the presence of microsatellite instability (MSI) and/or loss of expression of one or more of the DNA MMR proteins, MutL homolog 1 (MLH1), MutS homolog 2 (MSH2), MutS homolog 6 (MSH6) and post meiotic segregation increased 1 homolog 2 (PMS2). Identifying dMMR tumors is important for understanding disease prognosis,1 determining response to immune checkpoint inhibition therapy,2 and identifying patients with Lynch syndrome. Lynch syndrome is the most common inherited cancer predisposition disorder and, therefore, the Evaluation of Genomic Applications in Practice and Prevention Working Group recommends that all newly diagnosed colorectal cancers (CRCs) and endometrial cancers (ECs) are screened for dMMR to improve the identification of carriers.3,4
The dMMR mutator phenotype arises in tumors where errors occur during the DNA replication process.5 Specifically, defects in the components of the MMR system responsible for the recognition of mismatches, such as single-nucleotide variants (SNVs) and insertions-deletions (INDELs), can lead to the development of numerous frameshift mutations in coding and noncoding microsatellite regions.6 dMMR is related to biallelic inactivation of one of the MMR genes, resulting from either somatic methylation of the MLH1 gene promoter region7 or double somatic MMR gene mutations8 (sporadic dMMR), germline pathogenic variants in the MMR genes,9 or deletions in the 3′ end of the EPCAM gene10 (inherited dMMR). CRCs, ECs, and sebaceous skin tumors (SSTs), including sebaceous adenomas, carcinomas, and sebaceomas, are tissue types that demonstrate the highest frequencies of dMMR, where up to 26%,11 31%,11 and 31%12 of these tissue types, respectively, present with the dMMR phenotype, followed by stomach cancer at 19%.11
The most common approach for identifying dMMR tumors is by assessing MMR protein expression through immunohistochemistry (MMR IHC)13,14 and/or by testing for high levels of microsatellite instability using PCRs (MSI-PCR).15 Although both screening methods are commonly used, each presents advantages and limitations. The advantages of performing MMR IHC include simple experimental execution, short turnaround time, low associated costs, as well as giving an indication of the defective gene.16 However, false-positive or false-negative MMR IHC results can occur because of technical artifacts, variable performance of different MMR antibodies, and missense pathogenic variants causing falsely retained MMR protein expression and inherent variability in the interpretation of the staining by different pathologists.16,17 Further challenges include the interpretation of weaker staining in less proliferative tissue and heterogeneous patterns of MMR protein loss.18, 19, 20, 21, 22, 23, 24, 25
Although MMR IHC is more widely adopted in the clinical setting, MSI-PCR remains the gold standard for detecting dMMR16; to date, multiple markers have been identified to call MSI in tumor samples.26 The limitations for MSI-PCRs include additional laboratory implementation requirements related to tissue DNA extraction and increased labor costs; both can lead to a delay in receiving test results.16 Nonetheless, MMR IHC and MSI-PCR methods have proven to be effective for identifying dMMR in CRC samples,27 with a reported concordance of 91.9%,16 but the accuracy for either of these tools can decrease when applied to different tissue types.28 As next-generation sequencing (NGS) becomes more widely adopted for precision oncology, there is an increasing need to accurately determine tumor MMR status using NGS data.
To date, several tools have been developed to assess MSI from NGS data, including MSISensor,29 MSIseq,30 MANTIS,31 and, more recently, MSMuTect.32 To the best of our knowledge, the comparison of these four MSI tools on the same tumors has not yet been performed. In addition to MSI, other tumor features derived from NGS have been shown to be associated with dMMR, such as tumor mutational burden (TMB)33 and tumor mutational signatures (TMSs).34 TMB, characterized by high SNV and INDEL counts, is a biomarker for response to immune checkpoint inhibition therapy35,36 and is increased in dMMR tumors.37
TMSs aggregate tens to thousands of the observed somatic mutations within a tumor into patterns related to the underlying mutational processes.38,39 The predominant TMS framework, published on the Catalogue of Somatic Mutations in Cancer (COSMIC) website, defines 107 different signature definitions categorized into three distinct subgroups: i) 78 single-base substitutions (SBSs), where seven of the SBS signatures (SBS6, SBS14, SBS15, SBS20, SBS21, SBS26, and SBS44) are associated with dMMR; ii) 18 small (1 to 50 bp) insertions and deletions or ID signatures, where ID1, ID2, and ID7 are associated with dMMR; and iii) 11 doublet-base substitutions or DBS signatures, where DBS7 and DBS10 have both been previously associated with dMMR.34 However, DBS signatures have a reported low prevalence in CRC compared with other tissue types, so they were excluded from our study.39 Previously, we have shown that the combination of individual TMSs can improve the ability of TMSs to discriminate important molecular and genetic subtypes of CRC, including identifying germline biallelic carriers of pathogenic variants in the MUTYH gene by combining SBS18 and SBS36.40,41 It was further observed that the combination of ID2 with ID7 (TMS ID2 + ID7) was the most informative for differentiating dMMR from MMR-proficient (pMMR) CRCs among all possible TMS combinations.40 To date, the comparison of MSI calling tools, somatic mutation counts, and TMB and TMS tumor features for determining the dMMR status in CRC tumors has not yet been undertaken.
In this study, we assessed 104 tumor features derived from whole-exome sequencing (WES) (Table 1), consisting of the MSI prediction tools (MSMuTect, MANTIS, MSIseq, and MSISensor), TMS (78 SBS and 18 ID signatures), TMS ID2 + ID7, TMB, and individual SNV and INDEL somatic mutation counts for their accuracy in predicting dMMR status in 300 well-characterized CRCs. Second, we investigated whether a combination of these tumor features, using either a statistical model or a simple approach that added individual features together (additive feature combination), could improve the dMMR prediction accuracy in WES CRC tumors. Finally, we evaluated the effectiveness of the top performing tumor features from the WES analysis, individually and in combination, in an independent set of CRCs, ECs, and SSTs that had undergone targeted multigene panel sequencing for their dMMR prediction accuracy.
Table 1.
The Breakdown of the 104 Tumor Features Calculated from Next-Generation Sequencing Analysis Included in This Study
| Feature type | Count | Name | Reference |
|---|---|---|---|
| Total | N = 104 | ||
| MSI tools | N = 4 | MSISensor | Niu et al,29 2014 |
| MSIseq | Ni Huang et al,30 2015 | ||
| MANTIS | Kautto et al,31 2017 | ||
| MSMuTect | Maruvka et al,32 2017 | ||
| TMSs | N = 97 | SBS (n = 78) | Tate et al,34 2019 |
| ID (n = 18) | Tate et al,34 2019 | ||
| ID2 + ID7 | Georgeson et al,40 2021 | ||
| Somatic mutation counts | N = 3 | INDELs | |
| SNVs | |||
| TMB (SNVs + INDELs/Mb) | Cancer Genome Atlas Network,33 2012 |
The 104 tumor features can be categorized into three distinct groups: MSI tools, TMSs, and somatic mutation counts. These features have previously been shown to be associated with MSI/DNA mismatch repair status, as indicated by the provided references. The MSI group consists of four MSI tools (namely, MSISensor, MSIseq, MANTIS, and MSMuTect). TMSs consisted of 78 SBSs, 18 IDs, and TMS ID2 + ID7. The somatic mutation count consisted of the SNV count, larger INDEL count, and the TMB, which was calculated as the combination of SNV and INDEL counts per Mb.
ID, small insertion/deletion; INDEL, insertion/deletion; Mb, megabase; MSI, microsatellite instability; SBS, single-base substitution; SNV, single-nucleotide variant; TMB, tumor mutational burden; TMS, tumor mutational signature.
Materials and Methods
Study Cohort
The study population included men and women retrospectively identified from five studies where pMMR or dMMR status was determined by MMR IHC and where an etiology for dMMR status could be defined [namely, a sporadic etiology caused by tumor MLH1 methylation or double somatic MMR mutations, or an inherited etiology caused by a germline MMR gene pathogenic variant (Lynch syndrome)]. The breakdown of participants included in this study by their dMMR and pMMR status, tissue type, and WES or panel sequencing is shown in Figure 1:
-
1)
the ANGELS study (Applying Novel Genomic approaches to Early-onset and suspected Lynch Syndrome colorectal and endometrial cancers)40 recruited participants who were diagnosed with CRC or EC between 2014 and 2021 and who were referred from family cancer clinics across Australia (n = 79). All ANGELS study participants provided informed consent, and the study was approved by the University of Melbourne human research ethics committee (number 1750748) and institutional review boards at each family cancer clinic;
-
2)
CRC- or EC-affected participants from the ACCFR (Australasian Colorectal Cancer Family Registry) were selected from both population- and clinic-based recruitment (n = 139);
-
3)
CRC-affected participants from the OFCCR (Ontario Familial Colorectal Cancer Registry) were population-based patients (aged <50 years) recruited from the Cancer Care Ontario (Toronto, ON, Canada) (n = 53). Study participants from both the ACCFR and OFCCR were recruited between 1998 and 2008, and were included according to the recruitment policy and eligibility criteria previously described.42,43 Informed consent was obtained from all study participants, and the study protocol was approved by the institutional human ethics committee at both study sites;
-
4)
CRC-affected participants from the WEHI (Walter and Eliza Hall Institute of Medical Research) study were recruited from the Royal Melbourne Hospital (Parkville, VIC, Australia) and the Western Hospital Footscray (Footscray, VIC, Australia), between January 1, 1993, and December 31, 2009.40 All patients provided written informed consent. The study was approved by human research ethics committees at both sites (number 12/19) (n = 80);
-
5)
SST-affected participants from the MTS (Muir-Torre Syndrome) study were referred between July 2016 and September 2021 following clinical diagnostic MMR IHC testing by Sullivan Nicolaides Pathology service in Brisbane12 or by family cancer clinics in Australia. Informed consent was obtained from the study participants, and the study protocol was approved by the human research ethics committee from the University of Melbourne (number 1648355) and by the relevant institutional human ethics committees (n = 20).
Figure 1.
Overview of the study design. In total, 300 whole-exome sequenced (WES) colorectal cancers (CRCs), consisting of 91 DNA mismatch repair–deficient (dMMR) and 209 DNA mismatch repair–proficient (pMMR) tumors were analyzed. We investigated 104 tumor features for their ability to distinguish dMMR from pMMR tumors consisting of four microsatellite instability (MSI) tools, 97 tumor mutational signature (TMS) definitions, tumor mutational burden (TMB), calculated as mutations per megabase, somatic insertion/deletion (INDEL), and somatic single-nucleotide variant (SNV) counts. We performed a 10-fold cross-validation approach with 100 repeats to calculate the mean accuracy on the test data set. The top 10 ranked individual tumor features, a Lasso regression model, and an additive feature combination approach were tested to determine the benefit of combining tumor features to improve dMMR prediction. The findings from these three approaches were tested on an independent set of targeted panel-sequenced tumors of CRC, endometrial cancer (EC), and sebaceous skin tumor (SST) tissue types with reported mean accuracies. TMS small insertion/deletion (ID) 2, slippage during DNA replication of the replicated DNA strand; TMS ID7, defective DNA mismatch repair; TMS single-base substitution (SBS) 20, concurrent POLD1 mutations and defective DNA mismatch repair; TMS SBS54, possible sequencing artifact; possible contamination with germline variants; TMS SBS15, defective DNA mismatch repair, as described by Catalogue of Somatic Mutations in Cancer.34
Tumor Categorization
MMR IHC testing was performed on formalin-fixed, paraffin-embedded (FFPE) tissues for all four MMR proteins for the ACCFR and OFCCR, as previously described,43, 44, 45 and a subset of these tumors also underwent MSI-PCR testing, as previously described.46 MMR IHC testing for the ANGELS and MTS studies was part of routine clinical assessment in pathology laboratories across Australia, reported by the duty pathologist. Fresh-frozen tissue specimens from the WEHI study were assessed for MLH1, MSH2, and MSH6 MMR IHC and MSI-PCR tested using BAT25, BAT26, D5S346, D2S123, and D17S250 MSI markers. Germline MMR gene testing (as described in Buchanan et al44) and tumor MLH1 promoter methylation testing by MethyLight (as described in Buchanan et al47) were performed on all dMMR tumors showing loss of MLH1/PMS2 protein expression or sole PMS2 loss by IHC. Tumors were considered to have double somatic MMR mutations when they were found to have two pathogenic/likely pathogenic somatic mutations or a single somatic pathogenic/likely pathogenic mutation in combination with presence of loss of heterozygosity. Germline pathogenic variants and somatic MMR gene mutations were confirmed in WES and targeted panel sequencing data before analysis. Therefore, for each of the dMMR tumors included in this study, we could confirm an inherited or acquired cause for their respective pattern of MMR IHC protein loss. Concurrently, for the pMMR tumors, evidence of a germline MMR pathogenic variant or double MMR somatic mutation in these tumor samples was not found.
All tumors in the study were assigned to one of four categories based on dMMR or pMMR status determined from MMR IHC and/or MSI-PCR and based on the cause for dMMR:
-
1)
dMMR–Lynch syndrome (dMMR-LS): identified carrier of a germline pathogenic variant in one of the DNA MMR genes where the corresponding tumor showed commensurate loss of MMR protein expression by IHC;
-
2)
dMMR–MLH1 methylation (dMMR-MLH1me): tumors were positive for methylation of the MLH1 gene promoter C region48 and showed loss of MLH1 and PMS2 protein expression by IHC without a germline MMR gene pathogenic variant for tumors from the ANGELS, ACCFR, OFCCR, and MTS studies. For the WEHI study derived tumors, 8 of 15 of the MLH1me tumors had MLH1/PMS2 loss by IHC, whereas the remaining 7 of 15 dMMR-MLH1me tumors had only MSI-PCR performed and, therefore, loss of MLH1/PMS2 protein expression could not be confirmed;
-
3)
dMMR–double somatic: tumors harbored two somatic mutations (SNVs and/or loss of heterozygosity) in the same MMR gene that showed loss of protein expression by IHC with no identified pathogenic germline MMR gene variant; and
-
4)
pMMR: tumors showed normal expression of all four MMR proteins and did not show presence of double somatic MMR gene mutations or a germline MMR gene pathogenic variant.
The three dMMR subtypes (dMMR-LS, dMMR–double somatic, and dMMR-MLH1me) were combined as a single dMMR tumor group in downstream analysis.
Whole-Exome and Targeted Panel Sequencing Capture Regions
The targeted panel was based on the design described in Zaidi et al,49 consisting of probes targeting the following regions: i) 298 genes incorporating key hereditary CRC50, 51, 52 and EC53 risk genes and genes that are frequently mutated as identified by The Cancer Genome Atlas data,33,54,55 ii) 28 microsatellite loci, including the five gold standard MSI markers (BAT25, BAT26, NR-21, NR-24, and MONO-27) currently implemented in routine MSI-PCR diagnostics, iii) 212 homopolymer regions distributed genome-wide to assess for MSI in tumor samples, and iv) 56 copy number variants known to be susceptible to copy number changes in CRCs. The panel capture was 2.005 megabases (Mb) in size. The WES capture incorporates all exonic regions within the genome and is 67.296 Mb in size. The panel additionally included capture of intronic regions within the MMR genes, which the WES capture did not cover.
Next-Generation Sequencing
In total, 300 CRC tumors were sequenced by WES, and 71 tumors (29 CRCs, 22 ECs, and 20 SSTs) were sequenced by the targeted multigene panel (Figure 1). FFPE CRC, EC, or SST tissues were macrodissected and DNA extracted using the QIAamp DNA FFPE Tissue Kit (Qiagen, Hilden, Germany), according to the manufacturer's instructions. Peripheral blood–derived DNA was extracted using the DNeasy blood and tissue kit (Qiagen) and sequenced as germline references.
The WES capture was the Agilent Clinical Research Exome V2 kit (Agilent Technologies, Santa Clara, CA) with sequencing performed on an Illumina (San Diego, CA) NovaSeq 6000 comprising 150-bp paired-end reads performed at the Australian Genome Research Facility.40 For the WEHI CRCs, exome was enriched using the TruSeq Exome Enrichment Kit (Illumina) and 100-bp paired-end read sequencing performed on an Illumina HiSeq 2000 at the Australian Genome Research Facility.40 The on-target coverage for the 300 WES samples had a median of 323.7 for the FFPE tumor DNA samples and 137.4 for blood-derived DNA samples, with an interquartile range of 111.8 to 426.4 and 100.6 to 204.9, respectively.
Library preparation for targeted panel sequencing was performed using the SureSelect Low Input Target Enrichment System (Agilent Technologies) using standard protocol and sequenced on an Illumina NovaSeq 6000 comprising 150-bp paired-end reads performed at the Australian Genome Research Facility. The on-target coverage for the 71 panel-sequenced samples was (median and interquartile range) 919.3 and 694.6 to 1164.9 for FFPE tumor DNA samples and 160.6 and 135.8 to 178.0 for blood-derived DNA samples.
Bioinformatics Pipeline
For both WES and targeted panel sequenced samples, adapter sequences were trimmed from raw FASTQ files using Trimmomatic version 0.3856 and aligned to the GRCh37 human reference genome using Burrows-Wheeler Aligner version 0.7.12. Germline variants, somatic variants (SNVs), and somatic INDELs were called using Strelka version 2.9.2 (Illumina) using the recommended workflow.57 TMSs were calculated using the predefined set of 78 SBS and 18 ID signatures published on COSMIC as version 3.2 (COSMIC, https://cancer.sanger.ac.uk/signatures, last accessed June 15, 2022).34 Variants outside the WES and panel capture regions were excluded, and variants with the PASS filter called from Strelka were retained. Additional variant filters included were restrictions to a minimum depth of 50× for germline and tumor samples with a minimum variant allele frequency of 10%, as detailed previously.40
Selection of Features of Interest
The 104 tumor features selected for analysis in this study are shown in Table 1. Several tools have been developed to assess MSI from NGS data. Our analysis focused on MSMuTect version 2.0.5,32 MANTIS version 1.0.4,31 MSIseq version 1.0.0,30 and MSISensor version 0.5.29 Tumors were classified as having high levels of MSI or as microsatellite stable. All SBS (n = 78) and ID (n = 18) TMSs as described by COSMIC were assessed,34 but the DBS TMSs were excluded because of their reported low prevalence in CRCs.39 Combining ID2 and ID7 TMSs enables detection of dMMR CRCs40 and, therefore, they were included as a tumor feature in this study. Somatic mutation counts (namely, SNVs or INDELs), as well as TMB (SNV and INDEL mutation count combined/Mb) were each included, given previous associations with tumor dMMR status.58
Feature Performance Evaluation in WES Data from CRCs
One hundred and four tumor features calculated from WES from 209 pMMR CRCs and 91 dMMR CRCs (pMMR/dMMR ratio = 2.3:1) were assessed (Figure 1). The dMMR CRCs comprised dMMR-LS tumors (n = 49), dMMR-MLH1me tumors (n = 26), and dMMR–double somatic tumors (n = 16). All 300 CRCs were randomly partitioned into a training set (80% of CRCs) and a test set (20% of CRCs), while maintaining the same pMMR/dMMR ratio, using caret R package (https://cran.r-project.org/web/packages/caret/index.html). A 10-fold cross-validation approach was performed on the training set (repeated 100×) to calculate the average classification accuracy by fitting a generalized linear model and determining the error rate, specificity, sensitivity, and the area under the curve (AUC) with corresponding 95% CIs. On the basis of the unequal distribution of dMMR and pMMR tumors in the WES data set, the no information rate was 69.5%, indicating that any feature with this prediction accuracy was equivalent to selecting a dMMR sample by chance.
Tumor feature analysis of the WES CRC data set comprised three different approaches.
Individual Tumor Feature Assessment
Each of the 104 tumor features was assessed individually and then ranked by their accuracy in identifying dMMR tumors. Individual CRC tumor features with a prediction accuracy >80% from the WES data were considered good predictors for differentiating dMMR from pMMR tumors and were included in downstream analyses.
Generation of a Statistical Model by Combining Tumor Features
It was investigated whether combining tumor features using a Lasso penalized regression model59 could improve the overall dMMR prediction accuracy in CRC. Lasso enables the simultaneous parameter estimation and variable selection as well as having been shown to reduce overfitting when compared with conventional maximum likelihood regression models. Lasso regression has a tuning parameter called λ that controls which features are included in the regression model by shrinking the coefficient or weighting of individual features within the model toward 0, helping with the exclusion of some of the features from integration into the final model via a penalization process using cross-validation.
Applying an Additive Feature Combination Count
Our third approach investigated combining the top ranked individual tumor features in an additive approach (additive feature combination). Specifically, the tumor features that achieved a mean prediction accuracy >95% from the WES CRC analysis (from the individual tumor feature assessment) were included in this approach and added together to give an overall count. The bimodal distribution supported a majority vote decision on dMMR status.
Assessment of Individual Tumor Features, the Statistical Model, and Additive Feature Combination Approaches Derived from the WES Analysis on Panel-Sequenced CRCs, ECs, and SSTs
The top individual tumor features determined from best performing Lasso model and the additive feature combination approach were then assessed for their dMMR prediction accuracy in three independent tumor sets composed of n = 29 CRCs, n = 22 ECs, and n = 20 SSTs tested by targeted multigene panel sequencing. The no information rate for features analyzed from the panel data set was at 71.8%, indicating a prediction accuracy of this value was similar to selecting a dMMR sample by chance.
Statistical Analysis
All statistical analyses were performed using the R programming language version 4.1.0 (R Core Team, https://www.R-project.org, last accessed November 22, 2022). The tidyverse package version 1.3.160 was used for data import, tidying, and visualization purposes, and the caret version 6.0-9.0 package (https://cran.r-project.org/web/packages/caret/index.html) was used for cross-validation. Receiving operator curves were generated using the pROC package version 1.18.0,61 with the AUC being determined using the cvAUC package version 1.1.4 (https://cran.r-project.org/web/packages/cvAUC/index.html). Statistical models were fitted using the Lasso (glmnet version 4.1-3)62 package. The cutpointr version 1.1.1 package63 was used for estimation of the best cut points or thresholds, which maximize the Youden index (true-positive rate minus false-positive rate over all possible cut points), defined as the most optimal threshold in binary disease classification tasks. Herein, the cutpointr package determines a recommended threshold that best differentiates dMMR from pMMR cases for each feature and validates its performance using bootstrapping. The average weight for each group was calculated using the plyr version 1.0.7 package.64 The ggplot2 version 3.3.5 package65 was used for data visualization in combination with hrbrthemes version 0.8.0 (https://cran.r-project.org/web/packages/hrbrthemes/index.html) for histogram generation and ggrepel version 0.9.1 (https://cran.r-project.org/web/packages/ggrepel/index.html) for histogram annotations. Correlation scores between the dMMR and pMMR groups were estimated by a heteroscedastic two-tailed t-test. P < 0.05 was considered statistically significant. The 95% CIs for the WES data were calculated using the binomial (Clopper-Pearson) exact method66 and for the targeted panel data using the binom version 1.1-1 package in R (https://cran.r-project.org/web/packages/binom/index.html).
Data Availability Statement
The data generated during and/or analyzed during the current study are included in this published article (and its supplemental information files/source data file). These data are available from the Colon Cancer Family Registry via a request to collaborate with the Colon Cancer Family Registry application process approved by the NIH/National Cancer Institute (http://www.coloncfr.org/collaboration, last accessed October 21, 2022).
Results
The initial performance evaluation of 104 tumor features assessed 209 (69.7%) pMMR CRCs and 91 (30.3%) dMMR CRCs sequenced by WES (Supplemental Table S1). The clinicopathologic characteristics, pattern of MMR IHC loss, and dMMR etiology are summarized in Supplemental Table S2. The mean ± SD age at CRC diagnosis for the dMMR group was 51 ± 15.0 years, with 62.6% being female patients, and 49 ± 16.3 years, with 55.5% being female patients for the pMMR group. The values for each of the 104 tumor features derived from targeted panel sequencing for the 29 CRCs, 22 ECs, and 20 SSTs can be found in Supplemental Table S3. The clinicopathologic characteristics, pattern of MMR IHC loss, and dMMR etiology for these tumors are summarized in Supplemental Table S4. Within the panel-sequenced tumors, the proportion of dMMR for the CRC, EC, and SST subsets was 72.4% (21/29), 81.8% (18/22), and 65.0% (13/20), respectively. The predominant dMMR subtype across the CRC WES and targeted panel-sequenced tumors was dMMR-LS (53.8% and 66.7%, respectively). Within the dMMR subgroup, the most predominant pattern of loss observed in CRCs and ECs was MLH1/PMS2 (WES CRCs: 65.9%; panel CRCs: 47.6%; and ECs: 50.0%), whereas for the SST tumors, this was MSH2/MSH6 loss (76.9%). Tumors showing less common patterns of MMR loss, including solitary loss of MSH6 or PMS2 by IHC, were present in both the WES CRCs (16.5%) and panel-sequenced tumors (19.2%); however, sole PMS2 loss cases were absent from the EC and SST cohorts.
Assessment of Tumor Features for dMMR Prediction Accuracy in WES CRCs
Individual Tumor Feature Assessment
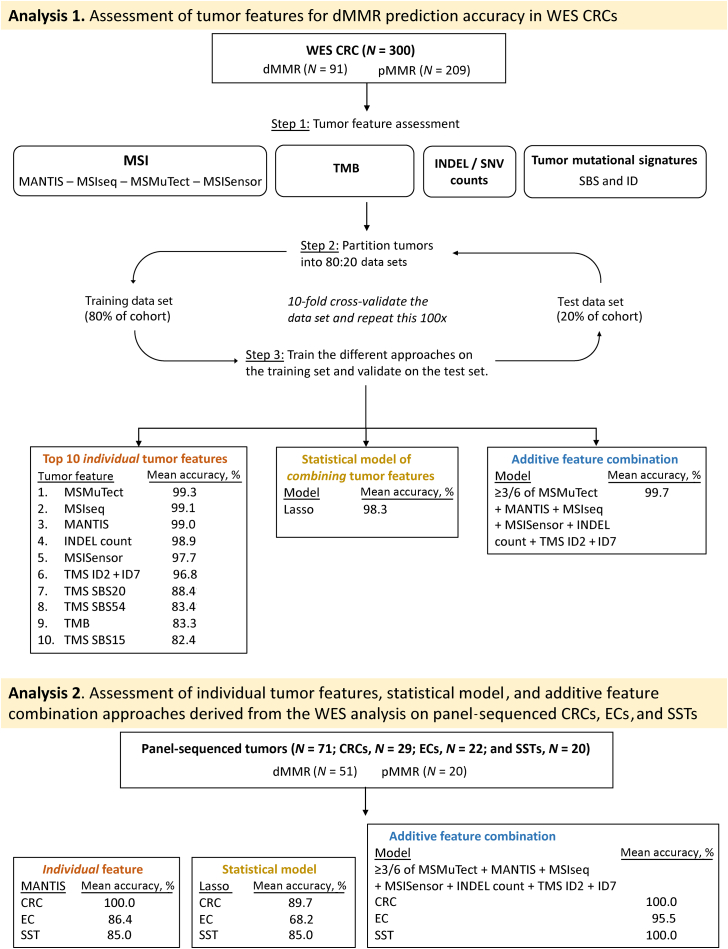
Twelve of the 104 tumor features derived from WES had a mean dMMR prediction accuracy >80% on the test data set (Table 2). The mean accuracy for the remaining 92 features is shown in Supplemental Table S5. The four MSI tools were among the best predictors, with MSMuTect, MSIseq, and MANTIS each achieving a mean prediction accuracy of ≥99.0%, with MSMuTect achieving the highest accuracy (99.3%; 95% CI, 99.1%–99.5%) (Table 2). The combination of TMS ID2 + ID7 achieved an accuracy of 96.8% (95% CI, 96.4%–97.2%) and outperformed these signatures individually (Table 2). To avoid collinearity issues between the combined TMS ID2 + ID7 variable with the individual TMS ID2 and TMS ID7 features, the latter were excluded from downstream analysis as they provided a lower prediction score. Therefore, the remaining 10 features were considered as the top 10 dMMR predictors and included in subsequent analyses (Figure 1).
Table 2.
Performance of the Top Tumor Features Demonstrating a Prediction Accuracy >80% Ranked by Highest Mean Accuracy from WES CRCs
| Tumor feature | Mean accuracy, % | Error rate, % | 95% CI for accuracy, % | Mean sensitivity, % | 95% CI for sensitivity, % | Mean specificity, % | 95% CI for specificity, % | Mean AUC, % | 95% CI for AUC, % |
|---|---|---|---|---|---|---|---|---|---|
| MSMuTect | 99.3 | 0.7 | 99.1–99.5 | 97.6 | 96.9–98.3 | 100.0 | NA | 98.8 | 98.5–99.1 |
| MSIseq | 99.1 | 0.9 | 98.9–99.4 | 97.7 | 97.0–98.3 | 99.8 | 99.6–100.0 | 98.7 | 98.4–99.1 |
| MANTIS | 99.0 | 1.0 | 98.8–99.2 | 97.1 | 96.4–97.7 | 99.9 | 99.8–100.0 | 98.5 | 98.1–98.8 |
| INDEL count | 98.9 | 1.1 | 98.7–99.2 | 97.7 | 97.0–98.3 | 99.5 | 99.2–99.8 | 98.6 | 98.2–98.9 |
| MSISensor | 97.7 | 2.3 | 97.3–98.0 | 93.4 | 92.4–94.5 | 99.5 | 99.3–99.7 | 96.5 | 96.0–97.0 |
| TMS ID2 + ID7 | 96.8 | 3.2 | 96.4–97.2 | 94.2 | 93.2–95.2 | 97.9 | 97.5–98.4 | 96.0 | 95.5–96.6 |
| TMS ID2 | 93.3 | 6.7 | 92.8–93.8 | 90.7 | 89.5–91.9 | 94.4 | 93.7–95.1 | 92.6 | 92.0–93.1 |
| TMS SBS20 | 88.4 | 11.6 | 87.6–89.2 | 68.9 | 66.6–71.2 | 97.0 | 96.4–97.6 | 82.9 | 81.8–84.1 |
| TMS ID7 | 87.6 | 12.4 | 87.0–88.3 | 74.2 | 72.6–75.9 | 93.5 | 92.8–94.2 | 83.9 | 83.0–84.7 |
| TMS SBS54 | 83.4 | 16.6 | 82.6–84.2 | 59.4 | 57.5–61.4 | 93.9 | 93.1–94.7 | 76.7 | 75.6–77.7 |
| TMB | 83.3 | 16.7 | 82.6–83.9 | 57.8 | 55.2–60.4 | 94.5 | 93.7–95.2 | 76.1 | 75.0–77.3 |
| TMS SBS15 | 82.4 | 17.6 | 81.5–83.3 | 58.8 | 56.5–61.1 | 92.8 | 91.9–93.7 | 75.8 | 74.6–77.0 |
The mean accuracy values after 10-fold cross-validation with 100 repeats, error rate, mean sensitivity, mean specificity, and mean AUCs with corresponding 95% CIs are shown for each of the top 10 predicting tumor features, MSMuTect, MSIseq, MANTIS, INDEL count, MSISensor, TMS ID2 + ID7, TMS ID2, TMS SBS20, TMS ID7, TMS SBS54, TMB, and TMS SBS15, from the WES CRC analysis.
AUC, area under the curve; CRC, colorectal cancer; ID, small insertion/deletion; INDEL, insertion/deletion; NA, not applicable; SBS, single-base substitutions; TMB, tumor mutational burden; TMS, tumor mutational signature; WES, whole-exome sequencing.
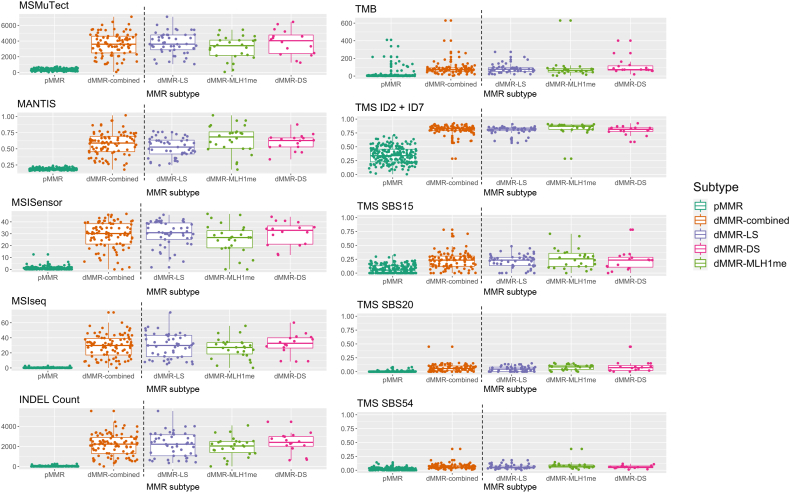
The mean, SD, and range of values for each of these top 10 dMMR predictive features by MMR status and by dMMR subtype for the 300 WES CRCs are shown in Supplemental Table S6. For each of these features, the mean values were significantly different between the dMMR and pMMR CRCs (all P <1 × 10−12 from a two-tailed t-test), with TMS ID2 + ID7 showing the most significant difference (P = 7.775 × 10−98), although MSISensor presented with the highest Cohen d effect size of 4.5, indicating that the means of the pMMR and dMMR groups differed by more than four times the SD (Supplemental Table S6). The variation in proportion or counts was larger in the dMMR tumors than in the pMMR tumors for all but one of these top 10 features where TMS ID2 + ID7 demonstrated a broad range of values in the pMMR CRCs compared with the dMMR CRCs (Figure 2 and Supplemental Table S6).
Figure 2.
Tumor distribution for each of the top 10 predicting features, MSMuTect, MANTIS, MSISensor, MSIseq, insertion/deletion (INDEL) count, tumor mutational burden (TMB; calculated as mutations/megabase), tumor mutational signature (TMS) small insertion/deletion (ID) 2 + ID7, TMS single-base substitution (SBS) 15, TMS SBS20, and TMS SBS54, as determined from the whole-exome sequencing analysis of colorectal cancers. The left of the vertical dotted line shows boxplots comparing the distribution of tumors by DNA mismatch repair (MMR) status: MMR proficient (pMMR) and MMR deficient (dMMR). The right of the vertical dotted line shows boxplots for each of the dMMR subgroups: dMMR–Lynch syndrome (LS), dMMR–double-somatic MMR gene mutation (DS), and dMMR–MLH1 promoter methylation (MLH1me).
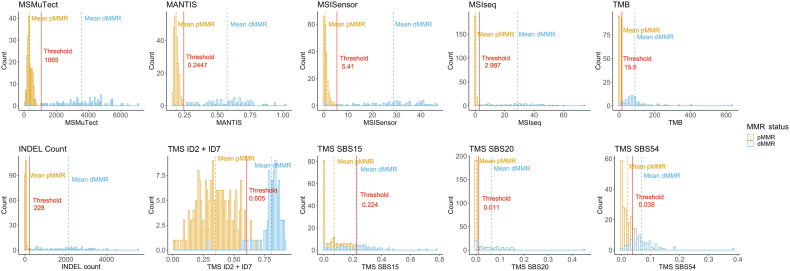
The AUCs for the top 10 features when taking all possible thresholds into account are shown in Supplemental Figure S1. The MSI prediction tools MSMuTect, MSIseq, and MANTIS as well as INDEL count demonstrated the best AUCs. In addition, the recommended thresholds for each feature were calculated for differentiating dMMR from pMMR CRCs using the method described in Materials and Methods (Supplemental Table S7). When applying these thresholds, it was not possible to achieve a complete separation between the dMMR and pMMR tumors for each of the tumor features (Figure 3).
Figure 3.
Determination of thresholds for differentiating DNA mismatch repair (MMR)–deficient (dMMR) from MMR-proficient (pMMR) colorectal cancers (CRCs) using whole-exome sequencing (WES) data for each of the top 10 performing tumor features. Bar graphs presenting the distribution of tumors after applying the recommended thresholds (red line) for each of the top 10 predicting tumor features, MSMuTect, MANTIS, MSISensor, MSIseq, insertion/deletion (INDEL) count, tumor mutational burden (TMB), tumor mutational signature (TMS) small insertion/deletion (ID) 2 + ID7, TMS single-base substitution (SBS) 15, TMS SBS20, and TMS SBS54, as determined from the WES CRC analysis. Orange indicates pMMR, and blue represents dMMR status.
Investigation of the CRCs misclassified on the basis of the individual tumor feature analysis demonstrated that the misclassification rate (error rate) for the MSI tools was low, with MSMuTect (2/300), MANTIS (1/300), MSIseq (1/300), and MSISensor (5/300) calling five or fewer incorrectly of 300 tumors (≤1.7% error rate). Of the CRCs misclassified by the MSI tools, only two tumors were misclassified by more than one MSI tool; both were dMMR-MLH1me CRCs classified as pMMR. Of note, one of these dMMR-MLH1me CRCs was misclassified as a pMMR tumor by 9 of the top 10 tumor features. The second misclassified dMMR-MLH1me CRC was classified as pMMR by MSMuTect and MSISensor but classified as dMMR by MSIseq and MANTIS (overall 6/10 features classified this CRC as dMMR). For INDEL count, 3 of 300 were incorrectly classified, where two pMMR CRCs were classified as dMMR. TMS ID2 + ID7 had 10 of 300 incorrect classifications, with seven pMMR tumors incorrectly called as dMMR. The remaining features from the top 10 prediction accuracy list demonstrated the following incorrect classifications: SBS20 (34/300), SBS54 (55/300), SBS15 (44/300), and TMB (19/300) encompassing incorrect calls in both directions (dMMR to pMMR and vice versa).
Generation of a Statistical Model by Combining Tumor Features
Combining features within a statistical model was assessed to determine if dMMR prediction accuracy could be improved. For this, a Lasso penalized logistic regression was performed. Herein, after calculating the best λ value, the combination of TMS ID2 + ID7 (coefficient = 5.29), MANTIS (coefficient = 1.70), and MSISensor (coefficient = 0.09) with SBS15 (coefficient = 2.25) provided the best prediction accuracy from all possible feature combinations, demonstrating a mean accuracy of 98.3% (95% CI, 0.981–0.986), sensitivity of 0.973 (95% CI, 0.966–0.980), and specificity of 1.000 (95% CI, 1.000–1.000) on the test set.
Assessing an Additive Feature Combination Count for dMMR Prediction
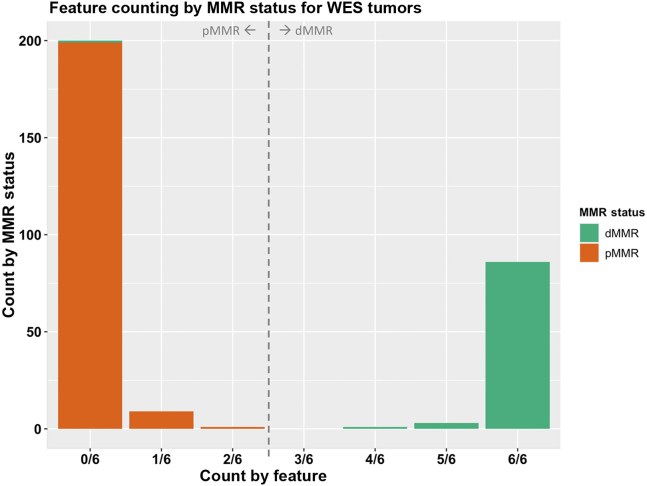
On the basis of the observation that the top performing tumor features from the individual feature analysis did not all misclassify the same CRCs, exploration was performed of a novel approach of combining tumor features together to increase the overall accuracy (ie, an additive tumor feature combination approach). This approach used a majority count of individual tumor features to overcome the small inaccuracies that each of the top tumor features displayed individually (ie, if one of these top dMMR predictive tumor features misclassified a CRC, then the other top dMMR predictive tumor features would correctly classify the same CRC and, thereby, achieve the correct classification overall). Six of the top 10 features from the 10-fold cross-validation analysis demonstrated a mean prediction accuracy of >95% and thus had the least number of incorrect CRC tumor classifications, consisting of MSMuTect, MANTIS, MSIseq, MSISensor, INDEL count, and TMS ID2 + ID7. The recommended threshold was applied for determining dMMR status determined previously for each tumor feature (Figure 3 and Supplemental Table S7) to derive a count of these six selected features, in which each feature is weighted equally. The results show a bimodal distribution across the 300 CRCs (Figure 4) where zero of six to two of six features correctly classified all the pMMR CRCs and four of six to six of six correctly classified all but one of the dMMR tumors with an accuracy of 99.7%. The only exception was the previously mentioned dMMR-MLH1me tumor, which did not meet the recommended thresholds for all six features and thus received a count of zero of six features, suggesting the CRC is pMMR rather than its initial dMMR status.
Figure 4.
The additive tumor feature combination approach demonstrating the distribution of counts of the top six tumor features by the DNA mismatch repair (MMR) status of the 300 colorectal cancers (CRCs) with whole-exome sequencing (WES). Bar graphs presenting the distribution of tumors after applying the additive tumor feature combination approach with the recommended thresholds from the WES CRC analysis using a count of three or more of the top six predictors from the WES CRC analysis, consisting of MSMuTect, MANTIS, MSIseq, MSISensor, insertion/deletion count, and tumor mutational signature small insertion/deletion (ID) 2 + ID7, for MMR status calling: MMR deficient (dMMR) versus MMR proficient (pMMR).
A summary of the results from the WES CRC analysis for the three approaches is shown in Table 3 and Figure 1.
Table 3.
Summary of the Best dMMR Prediction Results by Individual Tumor Feature, Lasso Regression Model, and the Additive Feature Combination Approach for the WES CRCs and the Panel-Sequenced CRCs, ECs, and SSTs
| WES | Performance of best individual feature |
Performance of statistical model |
Performance of additive feature combination approach |
|||
|---|---|---|---|---|---|---|
| Feature | Mean accuracy, % | Lasso | Mean accuracy, % | Feature combination | Mean accuracy, % | |
| CRC | MSMuTect | 99.3 | MANTIS + TMS ID2 + ID7 + MSISensor + TMS SBS15 | 98.3 | MSMuTect + MANTIS + MSIseq + MSISensor + INDEL count + TMS ID2 + ID7 | 99.7 |
| Panel | Feature | Accuracy, % | Lasso | Accuracy, % | Feature combination | Accuracy, % |
|---|---|---|---|---|---|---|
| CRC | MANTIS | 100.0 | MANTIS + TMS ID2 + ID7 + MSISensor + TMS SBS15 | 89.7 | MSMuTect + MANTIS + MSIseq + MSISensor + INDEL count + TMS ID2 + ID7 | 100.0 |
| EC | MANTIS | 86.4 | MANTIS + TMS ID2 + ID7 + MSISensor + TMS SBS15 | 68.2 | MSMuTect + MANTIS + MSIseq + MSISensor + INDEL count + TMS ID2 + ID7 | 95.5 |
| SST | MANTIS | 85.0 | MANTIS + TMS ID2 + ID7 + MSISensor + TMS SBS15 | 85.0 | MSMuTect + MANTIS + MSIseq + MSISensor + INDEL count + TMS ID2 + ID7 | 100.0 |
This table provides the top performing results from individual tumor feature, statistical model application (Lasso), and additive feature combination approach assessments for WES CRCs as well as targeted panel-sequenced CRCs, ECs, and SSTs.
CRC, colorectal cancer; dMMR, mismatch repair deficiency; EC, endometrial cancer; ID, small insertion/deletion; INDEL, insertion/deletion; SBS, single-base substitution; SST, sebaceous skin tumor; TMS, tumor mutational signature; WES, whole-exome sequencing.
Assessment of individual tumor features, Lasso statistical model, and additive feature combination approaches derived from the WES analysis on panel-sequenced CRCs, ECs, and SSTs.
To determine the generalizability of the findings from the three approaches performed on the WES CRCs, 71 tumors with targeted panel sequencing data were tested to evaluate performance on both a smaller capture and across different tissue types known to have a high prevalence of dMMR.
Evaluation of the Top Performing Individual Features from WES Analysis on the Panel-Sequenced CRCs, ECs, and SSTs
Of the top 10 dMMR tumor features from the WES CRC analysis, only four achieved a mean dMMR prediction accuracy of >80% in the panel-sequenced CRC tumors (Table 4). For ECs and SSTs, only one feature (MANTIS) and two features (MANTIS and TMS ID2 + ID7), respectively, of the top 10 tumor features achieved a mean dMMR prediction accuracy of >80% (Table 4). Across the three tissue types, MANTIS demonstrated the highest mean accuracy, achieving 100% (95% CI, 88.1%–100.0%) accuracy in the panel-sequenced CRCs, 86.4% accuracy in ECs (95% CI, 65.1%–97.1%), and 85% accuracy in SSTs (95% CI, 62.1%–96.8%) (Table 4). MSMuTect and INDEL count performed poorly in all three panel-sequenced tissue types compared with their accuracy in the WES CRCs. MSMuTect and INDEL count are features that provide absolute counts that in our data were two orders of magnitude smaller in the panel-sequenced tumors compared with the WES CRCs. The reduction in discriminatory ability is likely related to differences in the size (WES: 67.7 Mb; and panel: 2.0 Mb) and location (additional coverage of intronic regions of the MMR genes in the panel capture) of the regions covered by the WES and panel captures, resulting in a lower somatic mutation count.
Table 4.
Assessment of Top Performing Tumor Features from WES CRCs in Panel-Sequenced CRC, EC, and SST Test Sets
| Tumor feature | CRC, % |
EC, % |
SST, % |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Mean accuracy | 95% CI | Error rate | Mean accuracy | 95% CI | Error rate | Mean accuracy | 95% CI | Error rate | |
| MSMuTect | 27.6 | 12.7–47.2 | 72.4 | 18.2 | 5.2–40.3 | 81.8 | 35.0 | 15.4–59.2 | 65.0 |
| MSIseq | 82.8 | 64.2–94.2 | 17.2 | 68.2 | 45.1–86.1 | 31.8 | 65.0 | 40.8–84.6 | 35.0 |
| MANTIS | 100.0 | 88.1–100.0 | 0.0 | 86.4 | 65.1–97.1 | 13.6 | 85.0 | 62.1–96.8 | 15.0 |
| INDEL count | 27.6 | 12.7–47.2 | 72.4 | 18.2 | 5.2–40.3 | 81.8 | 35.0 | 15.4–59.2 | 65.0 |
| MSISensor | 96.6 | 82.2–99.9 | 3.4 | 77.3 | 54.6–92.2 | 22.7 | 75.0 | 50.9–91.3 | 25.0 |
| TMS ID2 + ID7 | 82.8 | 64.2–94.2 | 17.2 | 63.6 | 40.7–82.8 | 36.4 | 85.0 | 62.1–96.8 | 15.0 |
| TMS SBS20 | 69.0 | 49.2–84.7 | 31.0 | 50.0 | 28.2–71.8 | 50.0 | 40.0 | 19.1–63.9 | 60.0 |
| TMS SBS54 | 51.7 | 32.5–70.6 | 48.3 | 36.4 | 17.2–59.3 | 63.6 | 40.0 | 19.1–63.9 | 60.0 |
| TMB | 44.8 | 26.4–64.3 | 55.2 | 31.8 | 13.9–54.9 | 68.2 | 35.0 | 15.4–59.2 | 65.0 |
| TMS SBS15 | 44.8 | 26.4–64.3 | 55.2 | 27.3 | 10.7–50.2 | 72.7 | 60.0 | 36.1–80.9 | 40.0 |
Table presents the prediction accuracies, error rates, and corresponding 95% CIs for panel-sequenced CRCs, ECs, and SSTs for the top 10 predicting tumor features, MSMuTect, MSIseq, MANTIS, INDEL count, MSISensor, TMS ID2 + ID7, TMS SBS20, TMS SBS54, TMB (mutations/megabase), and TMS SBS15, from WES CRC analysis applied on panel-sequenced CRCs, ECs, and SSTs.
CRC, colorectal cancer; EC, endometrial cancer; ID, small insertion/deletion; INDEL, insertion/deletion; SBS, single-base substitution; SST, sebaceous skin tumor; TMB, tumor mutational burden; TMS, tumor mutational signature; WES, whole-exome sequencing.
The mean, SD, and range of values for each of these top 10 dMMR predicting features by MMR status and by dMMR subtype for each of CRC, EC, and SST tissue types are shown in Supplemental Table S8 and in Supplemental Figure S2, Supplemental Figure S3, and Supplemental Figure S4, respectively. The mean values of each of the top 10 predictors were significantly different between the dMMR and pMMR tumors in all three tissue types, except for TMS SBS15 in CRCs, MSISensor in ECs, TMB in ECs and SSTs, and TMS SBS20 and TMS SBS54 in SSTs. MSMuTect consistently had the highest Cohen d effect size of all top 10 tumor features for each tissue type, with the highest effect size observed in CRCs (3.2), indicating the mean of the dMMR and pMMR subgroups for this feature differ by approximately three SDs.
Evaluation of the Lasso Statistical Model on the Panel-Sequenced CRCs, ECs, and SSTs
From WES analysis, the Lasso statistical model, composed of TMS ID2 + ID7, MANTIS, MSISensor, and SBS15, achieved a mean prediction accuracy of 98.3%. When this model was applied, with the coefficients determined from the WES analysis, on these three independent panel-sequenced tissue types, the prediction accuracies were lower (CRC: 89.7%; EC: 68.2%; and SST: 85.0%) (Table 3).
Evaluation of the Additive Tumor Feature Combination Approach on the Panel-Sequenced CRCs, ECs, and SSTs
For each of the top 10 dMMR predictive tumor features, the optimal thresholds for the panel-sequenced CRCs, ECs, and SSTs were determined (Supplemental Table S7) and plotted by tissue type: CRC (Supplemental Figure S5), EC (Supplemental Figure S6), and SST (Supplemental Figure S7). The determined thresholds for MANTIS were consistent across both WES and panel captures as well as across tissue types, whereas the calculated thresholds for MSIseq were consistent for CRC across WES and panel captures but different to the thresholds determined for EC and SST. The remaining eight tumor features showed variability in their determined thresholds across both capture type and tissue type (Supplemental Table S7). As such, the thresholds determined for each tissue type for the panel-sequenced data were applied in the additive feature combination approach below.
The additive feature combination approach incorporates a count of MSMuTect, MANTIS, MSIseq, MSISensor, INDEL count, and TMS ID2 + ID7 tumor features to classify a tumor as dMMR. The distribution of the counts of these six tumor features determined for each tumor are shown for CRC (Supplemental Figure S8), EC (Supplemental Figure S9), and SSTs (Supplemental Figure S10). For each tissue type, all the dMMR tumors had three or more of six tumor features classify them as dMMR, except for a single dMMR-MLH1me EC (1/71, 1.4%), which scored zero of six and, therefore, was suggestive of pMMR status. This approach achieved accuracy scores of 100%, 95.5%, and 100% for CRC, EC, and SST, respectively (Table 3).
A summary of the WES CRC and CRC, EC, and SST panel-sequencing results for all three approaches is provided in Table 3.
Discussion
In this study, 104 tumor features calculated from next-generation sequencing data were compared for their accuracy in predicting dMMR status in 300 CRCs, 91 of which were dMMR determined by immunohistochemistry or MSI-PCR and with an established sporadic or inherited etiology for their dMMR status. Ten features achieved >80% dMMR prediction accuracy from the WES CRC tumors, with the highest accuracy predictors being the MSI tools MSMuTect, MSIseq, and MANTIS, all of which achieved ≥99% accuracy. The combination of TMS ID2 + ID7 achieved the highest mean accuracy for dMMR prediction of the 97 TMS features assessed. When applied to the targeted multigene panel setting, the performance of these 10 features was reduced not only in CRC but also for the EC and SSTs. In addition, we investigated two approaches that combined these top 10 performing tumor features to improve the overall prediction accuracy. The Lasso generated model achieved 98.3% accuracy in WES CRCs, although the performance of the model was reduced in the panel-sequenced CRCs, ECs, and SSTs. For both the WES CRCs and panel sequencing across tissue types, the additive tumor feature combination approach, where having three or more of the top six tumor features classify a tumor as dMMR, achieved the highest prediction accuracies of the three approaches tested.
To date, multiple tools to detect MSI from NGS data have been developed.67 NGS-based MSI tool development has been constantly evolving since the introduction of MSISensor29 and mSINGS,68 which were followed by MSIseq,30 MANTIS,31 and MSMuTect.32 However, to the best of our knowledge, neither a comparison of more than three MSI detection tools on the same tumor sample nor the effectiveness of these MSI tools specifically on SSTs has been performed to date. Previously, MANTIS has been compared with MSISensor, with the former showing superior sensitivity (97.18% versus 96.48%) and specificity (99.68% versus 98.73%).31 This was supported by our findings, and we additionally showed that across the WES and panel-tested CRCs, MANTIS provided the highest dMMR prediction accuracy and was shown to be the top performing feature in the ECs and SSTs as well. Recently, the US Food and Drug Administration approved MSISensor for detecting MSI in metastatic CRCs for selecting patients for immune checkpoint inhibition therapy.69 In our study, MSISensor had the lowest accuracy (97.7%) in WES CRCs of the four MSI tools tested, incorrectly classifying 5 of 300 CRCs. Seeking US Food and Drug Administration approval for other MSI tools in addition to MSISensor is warranted on the basis of our findings.
MSMuTect has been trained on 20 different tissue types using WES data and, therefore, it was not surprising it had the highest mean accuracy of the top performing tumor features in our WES CRC analysis. MSMuTect has been designed to accurately detect somatic MSI INDELs using a count of INDELs from the captured sequencing region.32 Thus, the MSI INDEL count from WES data (67.7 Mb) could be up to approximately 34× larger than that from panel data (2.0 Mb), which likely explains the poor performance of this tool observed in our panel-sequencing data test sets. When the MSMuTect threshold was adjusted for calling dMMR for panel data, MSMuTect showed improved discrimination of dMMR from pMMR tumors. This increase in prediction accuracy was also observed for the INDEL count, where adjusting the threshold for panel data improved the overall performance. Adjusting the threshold for panel-sequencing data enabled the inclusion of MSMuTect and INDEL count as two of the six tumor features in our additive feature combination approach that ultimately performed well on panel-sequenced tumors. Tumor features that calculate a percentage rather than raw counts, such as MANTIS, MSISensor, SBS TMS, and ID TMS, are more adaptable to changes in capture size. For example, our results showed that the calculated thresholds for differentiating dMMR from pMMR for MANTIS were consistent across both WES and panel captures as well as across tissue types. Therefore, training features that incorporate a count of genomic variants, such as INDELs, SNVs, and MSMuTect, on the capture size to improve dMMR prediction accuracy are recommended.
Although three ID TMSs (ID1, ID2, and ID7) are reported to be associated with dMMR,34 the results showed that the combination of ID2 and ID7 TMSs achieved the highest dMMR prediction accuracy of any of the TMS features in WES CRC tumors, outperforming ID2 or ID7 alone. Of the seven SBS TMSs that are associated with dMMR (SBS6, SBS14, SBS16, SBS20, SBS21, SBS26, and SBS44),34 only two, TMS SBS15 and TMS SBS20, showed >80% dMMR prediction accuracy in WES CRC tumors, but were shown to be poor predictors in the panel-sequenced tumors. Interestingly, TMS SBS54 was one of the top 10 dMMR predictors from the WES CRC analysis, although currently its proposed etiology in COSMIC is related to a “possible sequencing artifact and/or a possible contamination with germline variants.”34 Another study has shown that SBS15, SBS20, and SBS54 are observed in CRCs with a high immune cytolytic activity compared with cytolytic activity–low CRCs.70 Cytolytic activity–high CRCs have been shown to correlate with an increased somatic mutation load and high levels of MSI71; this may explain the observation of TMS SBS15, TMS SBS20, and TMS SBS54 demonstrating >80% dMMR prediction accuracy in the WES CRC analysis.
The combination of tumor features via the Lasso regression model achieved similar mean accuracy as the four MSI tools individually in the WES CRC analysis. The Lasso calculated final model that best distinguished dMMR from pMMR tumors in the WES CRC cohort consisted of TMS ID2 + ID7, MANTIS, MSISensor, and TMS SBS15. The statistical approach used to determine the final model assigns a weight (coefficient value) or confidence of how well each feature detects dMMR. As per generalized linear modeling method, the weight of any given feature is reduced as the model incorporates additional features. Hence, with MANTIS being one of the best predictors, its weighting was reduced when other features were added to the final model. This resulted in the Lasso model prediction accuracy being lower than MANTIS alone. Of note, because most of the approaches taken (ie, assessing features individually or in combination) already achieved a high prediction accuracy of approximately 99%, alternate modeling approaches, such as random forest, would not result in a significant improvement in dMMR prediction accuracy. The implementation of a dMMR prediction model, as proposed by the study findings, would require additional resources that could prove challenging in a practicing molecular pathology laboratory. As such, the results showed the MSI calling tools, in particular MANTIS, to be individually accurate across WES and panel-sequenced tumors should a combined six feature approach not be feasible.
Strengths of the study were a large sample of tumors, including dMMR tumors, with confirmed sporadic or inherited etiology concordant with MMR IHC and MSI-PCR results for both the WES and panel-sequenced data sets. Tumor MMR status combined with identified etiology provided a more reliable reference group of CRCs or truth set than would a group based on MMR IHC test results without etiological confirmation, given the known challenges that can lead to false-positive and false-negative MMR IHC results.16 Tumor features that can be readily derived from NGS data were assessed, ensuring that the findings have potential to be easily implemented in clinical diagnostics. The findings from the WES analysis were applied to panel data to determine the generalizability of the findings to smaller panel captures, such as those that are currently used in clinical diagnostics. The applicability of the study findings on different tissue types that display a high proportion of dMMR phenotype was demonstrated. The dMMR tumor samples included in this study were those showing the most frequent pattern of MMR IHC loss (namely, MLH1/PMS2 loss and MSH2/MSH6 loss) but also included tumors with solitary MSH6 loss or solitary PMS2 loss, ensuring a broad spectrum of dMMR patterns of loss were covered, which is particularly relevant given the identified challenges associated with interpretation of solitary MSH6 loss.72
There were several limitations of this study, including testing of only three tissue types. Testing of these tumor features and approaches in other tissue types, such as stomach cancer, which also has a high prevalence of dMMR overall and dMMR related to Lynch syndrome, would determine the suitability of these tumor features for inclusion in an additive feature combination approach in a pan-cancer setting. In addition, the sample size for the panel-sequenced tumors was limited for all three tissue types; however, there was a high proportion of dMMR in the tumors tested (72.4% for CRC, 81.8% for EC, and 65.0% for SST). It is well documented that germline MMR missense pathogenic variants can retain antigenicity, resulting in false-negative MMR IHC results.17 This study comprised 75 dMMR-LS cases, 21 (28%) were of missense variant type, where each demonstrated appropriate loss of the MMR protein by IHC (ie, no false negatives) and, therefore, the effectiveness of our approach on MMR missense variants that retain antigenicity could not be tested. Not all tumors were tested for both MMR IHC and MSI-PCR, meaning that the NGS-derived tumor features were compared largely with MMR IHC results and not MSI-PCR results. Although concordance between these two tests is typically not 100%,16 the 128 tumors there tested for both MMR IHC and MSI-PCR in this study did show 100% concordance. No tumor feature or approach achieved 100% accuracy in the CRC WES analysis. This was largely related to a single tumor (dMMR-MLH1me) from the WES CRC analysis that was called incorrectly by 9 of 10 top individual tumor features, suggesting the CRC was pMMR. Given this evidence, MLH1 methylation testing for this tumor was repeated using both MethyLight and methylation-sensitive high-resolution melting assays. Both assays found no evidence of MLH1 methylation in the tumor. This new MLH1 methylation result and the pMMR classification from our analysis suggest the initial dMMR classification was a false positive. If this CRC would initially have been categorized as a pMMR tumor, then MANTIS and MSIseq would have achieved 100% accuracy in the WES CRC analysis. Furthermore, the identification of an initial tumor misclassification provides strong support for evaluating multiple dMMR prediction tumor features and highlights the advantage of combining these features through an additive feature combination approach.
Conclusion
These findings provide an important comparison of tumor features for dMMR prediction, highlighting performance differences between capture size and tissue types. Our results demonstrate the high accuracy of multiple individual tumor features, including the MSI calling tools MSMuTect, MSIseq, MANTIS, and MSISensor, as well as INDEL count and the combination of TMS ID2 + ID7 for predicting dMMR status using WES CRCs. Moreover, the findings highlight the benefit of combining these six tumor features in a simple additive feature combination approach to improve dMMR prediction accuracy, particularly in targeted panel-sequencing data from CRCs, ECs, or SSTs. With the reported inaccuracies of MMR IHC and the increasing application of tumor sequencing for precision oncology, accurate NGS-derived dMMR detection has the potential to complement and even replace the current MMR IHC testing approach. Furthermore, revamping the current triaging approach to identify Lynch syndrome carriers from the sequential testing model to a one-stop tumor sequencing test that can accurately derive dMMR status and BRAFV600E mutation and identify germline and somatic MMR variants while providing information on therapeutic targets (eg, KRAS) and other hereditary cancer syndromes will have important implications for improving patient outcomes and cancer prevention.
Acknowledgments
We thank members of the Colorectal Oncogenomics Group and members from the Genomic Medicine and Family Cancer Clinic for support of this article; participants and staff from the Australasian and Ontario Colorectal Cancer Family Registries (ACCFR/OFCCR) and the ANGELS (Applying Novel Genomic approaches to Early-onset and suspected Lynch Syndrome colorectal and endometrial cancers), MTS (Muir-Torre Syndrome), and WEHI (Walter and Eliza Hall Institute of Medical Research) studies; especially Maggie Angelakos, Samantha Fox, and Allyson Templeton for supporting this study; the Australian Genome Research Facility for collaboration on this project; and A/Prof. Sue Finch (Melbourne Statistical Consulting Platform and Statistical Consulting Center at the University of Melbourne) for guidance with the statistical aspects of this study.
Footnotes
Supported by a National Health and Medical Research Council of Australia (NHMRC) project grant GNT1125269 (principal investigator D.D.B.), which supported the design, analysis, and interpretation of data; the Margaret and Irene Stewardson Fund Scholarship (R.W.); the Melbourne Research Scholarship (R.W.); NHMRC Investigator grants GNT1194896 (D.D.B.), GNT1195099 (M.A.J.), and GNT1194392 (A.K.W.); University of Melbourne Dame Kate Campbell Fellowship (D.D.B. and J.L.H.); the University of Melbourne Research Scholarship (P.G.); an NHMRC Senior Research Fellowship GNT1136119 (O.M.S.); a Canadian Institutes of Health Research Post-doctoral Fellowship (D.E.O.); and a Victorian Health and Medical Research Fellowship from the Victorian Government (B.J.P.).
Disclosures: None declared.
Supplemental material for this article can be found at http://doi.org/10.1016/j.jmoldx.2022.10.003.
Supplemental Data
The cross-validated areas under the curves (AUCs) for the top 10 performing tumor features to predict the DNA mismatch repair status in the whole-exome sequenced (WES) colorectal cancer (CRC) data set. Receiving operator curves presenting the AUCs for the top 10 performing tumor features, MSMuTect, MANTIS, MSISensor, MSIseq, insertion/deletion (INDEL) count, tumor mutational burden (TMB; calculated as mutations per megabase), tumor mutational signature (TMS) small insertion/deletion (ID) 2 + ID7, TMS single-base substitution (SBS) 15, TMS SBS20, and TMS SBS54, as determined from the WES CRC analysis. The 95% CIs for each feature are shown in red highlighting.
Tumor distribution for each of the top 10 predicting features, MSMuTect, MANTIS, MSISensor, MSIseq, insertion/deletion (INDEL) count, tumor mutational burden (TMB; calculated as mutations per megabase), tumor mutational signature (TMS) small insertion/deletion (ID) 2 + ID7, TMS single-base substitution (SBS) 15, TMS SBS20, and TMS SBS54, identified from the whole-exome sequencing colorectal cancer (CRC) analysis that were applied to targeted panel-sequenced CRCs. The left of the vertical dotted line shows boxplots comparing the distribution of tumors by DNA mismatch repair (MMR) status: MMR proficient (pMMR) and MMR deficient (dMMR). The right of the vertical dotted line shows boxplots for each of the dMMR subgroups: dMMR–Lynch syndrome (LS), dMMR–double-somatic MMR gene mutation (DS), and dMMR–MLH1 promoter methylation (MLH1me).
Tumor distribution for each of the top 10 predicting features, MSMuTect, MANTIS, MSISensor, MSIseq, insertion/deletion (INDEL) count, tumor mutational burden (TMB; calculated as mutations per megabase), tumor mutational signature (TMS) small insertion/deletion (ID) 2 + ID7, TMS single-base substitution (SBS) 15, TMS SBS20, and TMS SBS54, identified from the whole-exome sequencing colorectal cancer analysis that were applied to targeted panel-sequenced endometrial cancers. The left of the vertical dotted line shows boxplots comparing the distribution of tumors by DNA mismatch repair (MMR) status: MMR proficient (pMMR) and MMR deficient (dMMR). The right of the vertical dotted line shows boxplots for each of the dMMR subgroups: dMMR–Lynch syndrome (LS), dMMR–double-somatic MMR gene mutation (DS), and dMMR–MLH1 promoter methylation (MLH1me).
Tumor distribution for each of the top 10 predicting features, MSMuTect, MANTIS, MSISensor, MSIseq, insertion/deletion (INDEL) count, tumor mutational burden (TMB; calculated as mutations per megabase), tumor mutational signature (TMS) small insertion/deletion (ID) 2 + ID7, TMS single-base substitution (SBS) 15, TMS SBS20, and TMS SBS54, identified from the whole-exome sequencing colorectal cancer analysis that were applied to targeted panel-sequenced sebaceous skin tumors. The left of the vertical dotted line shows boxplots comparing the distribution of tumors by DNA mismatch repair (MMR) status: MMR proficient (pMMR) and MMR deficient (dMMR). The right of the vertical dotted line shows boxplots for each of the dMMR subgroups: dMMR–Lynch syndrome (LS), dMMR–double-somatic MMR gene mutation (DS), and dMMR–MLH1 promoter methylation (MLH1me).
Determination of the thresholds for the top 10 performing features for panel colorectal cancers (CRCs). Bar graphs presenting the distribution of tumors after applying the recommended thresholds for each of the top 10 predicting tumor features, MSMuTect, MANTIS, MSISensor, MSIseq, insertion/deletion (INDEL) count, tumor mutational burden (TMB; calculated as mutations per megabase), tumor mutational signature (TMS) small insertion/deletion (ID) 2 + ID7, TMS single-base substitution (SBS) 15, TMS SBS20, and TMS SBS54, for targeted panel-sequenced CRCs. Orange indicates DNA mismatch repair (MMR) proficient (pMMR), and blue represents MMR-deficient (dMMR) status.
Determination of the thresholds for the top 10 performing features for panel endometrial cancers (ECs). Bar graphs presenting the distribution of tumors after applying the recommended thresholds for each of the top 10 predicting tumor features, MSMuTect, MANTIS, MSISensor, MSIseq, insertion/deletion (INDEL) count, tumor mutational burden (TMB; calculated as mutations per megabase), tumor mutational signature (TMS) small insertion/deletion (ID) 2 + ID7, TMS single-base substitution (SBS) 15, TMS SBS20, and TMS SBS54, for targeted panel-sequenced ECs. Orange indicates DNA mismatch repair (MMR) proficient (pMMR), and blue represents MMR-deficient (dMMR) status.
Determination of the thresholds for the top 10 performing features for panel sebaceous skin tumors (SSTs). Bar graphs presenting the distribution of tumors after applying the recommended thresholds for each of the top 10 predicting tumor features, MSMuTect, MANTIS, MSISensor, MSIseq, insertion/deletion (INDEL) count, tumor mutational burden (TMB; calculated as mutations per megabase), tumor mutational signature (TMS) small insertion/deletion (ID) 2 + ID7, TMS single-base substitution (SBS) 15, TMS SBS20, and TMS SBS54, for targeted panel-sequenced SSTs. Orange indicates DNA mismatch repair (MMR) proficient (pMMR), and blue represents MMR-deficient (dMMR) status.
The additive tumor feature combination approach demonstrating the distribution of counts of the top six tumor features by the DNA mismatch repair (MMR) status of the 29 panel-sequenced colorectal cancers (CRCs). Bar graphs presenting the distribution of tumors after applying the additive tumor feature combination approach with the recommended thresholds for targeted panel-sequenced CRCs using a count of three or more of the top six predictors with >95% accuracy, consisting of MSMuTect, MANTIS, MSIseq, MSISensor, insertion/deletion count, and tumor mutational signature small insertion/deletion (ID) 2 + ID7 for MMR status calling: MMR deficient (dMMR) versus MMR proficient (pMMR).
The additive tumor feature combination approach demonstrating the distribution of counts of the top six tumor features by the DNA mismatch repair (MMR) status of the 22 panel-sequenced endometrial cancers (ECs). Bar graphs presenting the distribution of tumors after applying the additive tumor feature combination approach with the recommended thresholds for targeted panel-sequenced ECs using a count of three or more of the top six predictors with >95% accuracy, consisting of MSMuTect, MANTIS, MSIseq, MSISensor, insertion/deletion count, and tumor mutational signature small insertion/deletion (ID) 2 + ID7 for MMR status calling: MMR deficient (dMMR) versus MMR proficient (pMMR).
The additive tumor feature combination approach demonstrating the distribution of counts of the top six tumor features by the DNA mismatch repair (MMR) status of the 20 panel-sequenced sebaceous skin tumors (SSTs). Bar graphs presenting the distribution of tumors after applying the additive tumor feature combination approach with the recommended thresholds for targeted panel-sequenced SSTs using a count of three or more of the top six predictors with >95% accuracy, consisting of MSMuTect, MANTIS, MSIseq, MSISensor, insertion/deletion count, and tumor mutational signature small insertion/deletion (ID) 2 + ID7 for MMR status calling: MMR deficient (dMMR) versus MMR proficient (pMMR).
References
- 1.Gryfe R., Kim H., Hsieh E.T., Aronson M.D., Holowaty E.J., Bull S.B., Redston M., Gallinger S. Tumor microsatellite instability and clinical outcome in young patients with colorectal cancer. N Engl J Med. 2000;342:69–77. doi: 10.1056/NEJM200001133420201. [DOI] [PubMed] [Google Scholar]
- 2.Le D.T., Uram J.N., Wang H., Bartlett B.R., Kemberling H., Eyring A.D., Skora A.D., Luber B.S., Azad N.S., Laheru D., Biedrzycki B., Donehower R.C., Zaheer A., Fisher G.A., Crocenzi T.S., Lee J.J., Duffy S.M., Goldberg R.M., de la Chapelle A., Koshiji M., Bhaijee F., Huebner T., Hruban R.H., Wood L.D., Cuka N., Pardoll D.M., Papadopoulos N., Kinzler K.W., Zhou S., Cornish T.C., Taube J.M., Anders R.A., Eshleman J.R., Vogelstein B., Diaz L.A. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372:2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Recommendations from the EGAPP Working Group: genetic testing strategies in newly diagnosed individuals with colorectal cancer aimed at reducing morbidity and mortality from Lynch syndrome in relatives. Genet Med. 2009;11:35–41. doi: 10.1097/GIM.0b013e31818fa2ff. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Green R.F., Ari M., Kolor K., Dotson W.D., Bowen S., Habarta N., Rodriguez J.L., Richardson L.C., Khoury M.J. Evaluating the role of public health in implementation of genomics-related recommendations: a case study of hereditary cancers using the CDC Science Impact Framework. Genet Med. 2019;21:28–37. doi: 10.1038/s41436-018-0028-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baretti M., Le D.T. DNA mismatch repair in cancer. Pharmacol Ther. 2018;189:45–62. doi: 10.1016/j.pharmthera.2018.04.004. [DOI] [PubMed] [Google Scholar]
- 6.Eshleman J.R., Markowitz S.D. Mismatch repair defects in human carcinogenesis. Hum Mol Genet. 1996;5(Spec No):1489–1494. doi: 10.1093/hmg/5.supplement_1.1489. [DOI] [PubMed] [Google Scholar]
- 7.Young J., Simms L.A., Biden K.G., Wynter C., Whitehall V., Karamatic R., George J., Goldblatt J., Walpole I., Robin S.-A., Borten M.M., Stitz R., Searle J., McKeone D., Fraser L., Purdie D.R., Podger K., Price R., Buttenshaw R., Walsh M.D., Barker M., Leggett B.A., Jass J.R. Features of colorectal cancers with high-level microsatellite instability occurring in familial and sporadic settings. Am J Pathol. 2001;159:2107–2116. doi: 10.1016/S0002-9440(10)63062-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia-Closas M., Egan K.M., Abruzzo J., Newcomb P.A., Titus-Ernstoff L., Franklin T., Bender P.K., Beck J.C., Le Marchand L., Lum A., Alavanja M., Hayes R.B., Rutter J., Buetow K., Brinton L.A., Rothman N. Collection of genomic DNA from adults in epidemiological studies by buccal cytobrush and mouthwash. Cancer Epidemiol Biomarkers Prev. 2001;10:687–696. [PubMed] [Google Scholar]
- 9.Lynch H.T., Lynch P.M., Lanspa S.J., Snyder C.L., Lynch J.F., Boland C.R. Review of the Lynch syndrome: history, molecular genetics, screening, differential diagnosis, and medicolegal ramifications. Clin Genet. 2009;76:1–18. doi: 10.1111/j.1399-0004.2009.01230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ligtenberg M.J., Kuiper R.P., Chan T.L., Goossens M., Hebeda K.M., Voorendt M., Lee T.Y., Bodmer D., Hoenselaar E., Hendriks-Cornelissen S.J., Tsui W.Y., Kong C.K., Brunner H.G., van Kessel A.G., Yuen S.T., van Krieken J.H., Leung S.Y., Hoogerbrugge N. Heritable somatic methylation and inactivation of MSH2 in families with Lynch syndrome due to deletion of the 3’ exons of TACSTD1. Nat Genet. 2009;41:112–117. doi: 10.1038/ng.283. [DOI] [PubMed] [Google Scholar]
- 11.Bonneville R., Krook M.A., Kautto E.A., Miya J., Wing M.R., Chen H.-Z., Reeser J.W., Yu L., Roychowdhury S. Landscape of microsatellite instability across 39 cancer types. JCO Precis Oncol. 2017;2017 doi: 10.1200/PO.17.00073. PO.17.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walsh M.D., Jayasekara H., Huang A., Winship I.M., Buchanan D.D. Clinico-pathological predictors of mismatch repair deficiency in sebaceous neoplasia: a large case series from a single Australian private pathology service. Australas J Dermatol. 2019;60:126–133. doi: 10.1111/ajd.12958. [DOI] [PubMed] [Google Scholar]
- 13.Mascarenhas L., Shanley S., Mitchell G., Spurdle A.B., Macrae F., Pachter N., Buchanan D.D., Ward R.L., Fox S., Duxbury E., Driessen R., Boussioutas A. Current mismatch repair deficiency tumor testing practices and capabilities: a survey of Australian pathology providers. Asia Pac J Clin Oncol. 2018;14:417–425. doi: 10.1111/ajco.13076. [DOI] [PubMed] [Google Scholar]
- 14.Shia J. Immunohistochemistry versus microsatellite instability testing for screening colorectal cancer patients at risk for hereditary nonpolyposis colorectal cancer syndrome, part I: the utility of immunohistochemistry. J Mol Diagn. 2008;10:293–300. doi: 10.2353/jmoldx.2008.080031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boland C.R., Thibodeau S.N., Hamilton S.R., Sidransky D., Eshleman J.R., Burt R.W., Meltzer S.J., Rodriguez-Bigas M.A., Fodde R., Ranzani G.N., Srivastava S. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–5257. [PubMed] [Google Scholar]
- 16.Chen M.-L., Chen J.-Y., Hu J., Chen Q., Yu L.-X., Liu B.-R., Qian X.-P., Yang M. Comparison of microsatellite status detection methods in colorectal carcinoma. Int J Clin Exp Pathol. 2018;11:1431–1438. [PMC free article] [PubMed] [Google Scholar]
- 17.Rosty C., Clendenning M., Walsh M.D., Eriksen S.V., Southey M.C., Winship I.M., Macrae F.A., Boussioutas A., Poplawski N.K., Parry S., Arnold J., Young J.P., Casey G., Haile R.W., Gallinger S., Le Marchand L., Newcomb P.A., Potter J.D., DeRycke M., Lindor N.M., Thibodeau S.N., Baron J.A., Win A.K., Hopper J.L., Jenkins M.A., Buchanan D.D. Germline mutations in PMS2 and MLH1 in individuals with solitary loss of PMS2 expression in colorectal carcinomas from the Colon Cancer Family Registry Cohort. BMJ Open. 2016;6:e010293. doi: 10.1136/bmjopen-2015-010293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chapusot C., Martin L., Bouvier A.M., Bonithon-Kopp C., Ecarnot-Laubriet A., Rageot D., Ponnelle T., Laurent Puig P., Faivre J., Piard F. Microsatellite instability and intratumoural heterogeneity in 100 right-sided sporadic colon carcinomas. Br J Cancer. 2002;87:400–404. doi: 10.1038/sj.bjc.6600474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graham R.P., Kerr S.E., Butz M.L., Thibodeau S.N., Halling K.C., Smyrk T.C., Dina M.A., Waugh V.M., Rumilla K.M. Heterogenous MSH6 loss is a result of microsatellite instability within MSH6 and occurs in sporadic and hereditary colorectal and endometrial carcinomas. Am J Surg Pathol. 2015;39:1370–1376. doi: 10.1097/PAS.0000000000000459. [DOI] [PubMed] [Google Scholar]
- 20.Joost P., Veurink N., Holck S., Klarskov L., Bojesen A., Harbo M., Baldetorp B., Rambech E., Nilbert M. Heterogenous mismatch-repair status in colorectal cancer. Diagn Pathol. 2014;9:126. doi: 10.1186/1746-1596-9-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCarthy A.J., Capo-Chichi J.-M., Spence T., Grenier S., Stockley T., Kamel-Reid S., Serra S., Sabatini P., Chetty R. Heterogenous loss of mismatch repair (MMR) protein expression: a challenge for immunohistochemical interpretation and microsatellite instability (MSI) evaluation. J Pathol Clin Res. 2019;5:115–129. doi: 10.1002/cjp2.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pai R.K., Plesec T.P., Abdul-Karim F.W., Yang B., Marquard J., Shadrach B., Roma A.R. Abrupt loss of MLH1 and PMS2 expression in endometrial carcinoma: molecular and morphologic analysis of 6 cases. Am J Surg Pathol. 2015;39:993–999. doi: 10.1097/PAS.0000000000000415. [DOI] [PubMed] [Google Scholar]
- 23.Shia J., Zhang L., Shike M., Guo M., Stadler Z., Xiong X., Tang L.H., Vakiani E., Katabi N., Wang H., Bacares R., Ruggeri J., Boland C.R., Ladanyi M., Klimstra D.S. Secondary mutation in a coding mononucleotide tract in MSH6 causes loss of immunoexpression of MSH6 in colorectal carcinomas with MLH1/PMS2 deficiency. Mod Pathol. 2013;26:131–138. doi: 10.1038/modpathol.2012.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watkins J.C., Nucci M.R., Ritterhouse L.L., Howitt B.E., Sholl L.M. Unusual mismatch repair immunohistochemical patterns in endometrial carcinoma. Am J Surg Pathol. 2016;40:909–916. doi: 10.1097/PAS.0000000000000663. [DOI] [PubMed] [Google Scholar]
- 25.Watson N., Grieu F., Morris M., Harvey J., Stewart C., Schofield L., Goldblatt J., Iacopetta B. Heterogeneous staining for mismatch repair proteins during population-based prescreening for hereditary nonpolyposis colorectal cancer. J Mol Diagn. 2007;9:472–478. doi: 10.2353/jmoldx.2007.060162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baudrin L.G., Deleuze J.-F., How-Kit A. Molecular and computational methods for the detection of microsatellite instability in cancer. Front Oncol. 2018;8:621. doi: 10.3389/fonc.2018.00621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vasen H.F.A., Hendriks Y., de Jong A.E., van Puijenbroek M., Tops C., Bröcker-Vriends A.H.J.T., Wijnen JTh, Morreau H. Identification of HNPCC by molecular analysis of colorectal and endometrial tumors. Dis Markers. 2004;20:207–213. doi: 10.1155/2004/391039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siemanowski J., Schömig-Markiefka B., Buhl T., Haak A., Siebolts U., Dietmaier W., Arens N., Pauly N., Ataseven B., Büttner R., Merkelbach-Bruse S. Managing difficulties of microsatellite instability testing in endometrial cancer-limitations and advantages of four different PCR-based approaches. Cancers (Basel) 2021;13:1268. doi: 10.3390/cancers13061268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niu B., Ye K., Zhang Q., Lu C., Xie M., McLellan M.D., Wendl M.C., Ding L. MSIsensor: microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics. 2014;30:1015–1016. doi: 10.1093/bioinformatics/btt755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ni Huang M., McPherson J.R., Cutcutache I., Teh B.T., Tan P., Rozen S.G. MSIseq: software for assessing microsatellite instability from catalogs of somatic mutations. Sci Rep. 2015;5:13321. doi: 10.1038/srep13321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kautto E.A., Bonneville R., Miya J., Yu L., Krook M.A., Reeser J.W., Roychowdhury S. Performance evaluation for rapid detection of pan-cancer microsatellite instability with MANTIS. Oncotarget. 2017;8:7452–7463. doi: 10.18632/oncotarget.13918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maruvka Y.E., Mouw K.W., Karlic R., Parasuraman P., Kamburov A., Polak P., Haradhvala N.J., Hess J.M., Rheinbay E., Brody Y., Koren A., Braunstein L.Z., D’Andrea A., Lawrence M.S., Bass A., Bernards A., Michor F., Getz G. Analysis of somatic microsatellite indels identifies driver events in human tumors. Nat Biotechnol. 2017;35:951–959. doi: 10.1038/nbt.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tate J.G., Bamford S., Jubb H.C., Sondka Z., Beare D.M., Bindal N., Boutselakis H., Cole C.G., Creatore C., Dawson E., Fish P., Harsha B., Hathaway C., Jupe S.C., Kok C.Y., Noble K., Ponting L., Ramshaw C.C., Rye C.E., Speedy H.E., Stefancsik R., Thompson S.L., Wang S., Ward S., Campbell P.J., Forbes S.A. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res. 2019;47:D941–D947. doi: 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Panda A., Betigeri A., Subramanian K., Ross J.S., Pavlick D.C., Ali S., Markowski P., Silk A., Kaufman H.L., Lattime E., Mehnert J.M., Sullivan R., Lovly C.M., Sosman J., Johnson D.B., Bhanot G., Ganesan S. Identifying a clinically applicable mutational burden threshold as a potential biomarker of response to immune checkpoint therapy in solid tumors. JCO Precis Oncol. 2017;2017 doi: 10.1200/PO.17.00146. PO.17.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng M. Tumor mutation burden for predicting immune checkpoint blockade response: the more, the better. J Immunother Cancer. 2022;10:e003087. doi: 10.1136/jitc-2021-003087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang H., Sasson A., Srinivasan S., Golhar R., Greenawalt D.M., Geese W.J., Green G., Zerba K., Kirov S., Szustakowski J. Bioinformatic methods and bridging of assay results for reliable tumor mutational burden assessment in non-small-cell lung cancer. Mol Diagn Ther. 2019;23:507–520. doi: 10.1007/s40291-019-00408-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alexandrov L.B., Nik-Zainal S., Wedge D.C., Aparicio S.A.J.R., Behjati S., Biankin A.V., et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alexandrov L.B., Kim J., Haradhvala N.J., Huang M.N., Tian Ng A.W., Wu Y., Boot A., Covington K.R., Gordenin D.A., Bergstrom E.N., Islam S.M.A., Lopez-Bigas N., Klimczak L.J., McPherson J.R., Morganella S., Sabarinathan R., Wheeler D.A., Mustonen V., Getz G., Rozen S.G., Stratton M.R. The repertoire of mutational signatures in human cancer. Nature. 2020;578:94–101. doi: 10.1038/s41586-020-1943-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Georgeson P., Pope B.J., Rosty C., Clendenning M., Mahmood K., Joo J.E., Walker R., Hutchinson R.A., Preston S., Como J., Joseland S., Win A.K., Macrae F.A., Hopper J.L., Mouradov D., Gibbs P., Sieber O.M., O'Sullivan D.E., Brenner D.R., Gallinger S., Jenkins M.A., Winship I.M., Buchanan D.D. Evaluating the utility of tumour mutational signatures for identifying hereditary colorectal cancer and polyposis syndrome carriers. Gut. 2021;70:2138–2149. doi: 10.1136/gutjnl-2019-320462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Georgeson P., Harrison T.A., Pope B.J., Zaidi S.H., Qu C., Steinfelder R.S., et al. Identifying colorectal cancer caused by biallelic MUTYH pathogenic variants using tumor mutational signatures. Nat Commun. 2022;13:3254. doi: 10.1038/s41467-022-30916-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jenkins M.A., Win A.K., Templeton A.S., Angelakos M.S., Buchanan D.D., Cotterchio M., Figueiredo J.C., Thibodeau S.N., Baron J.A., Potter J.D., Hopper J.L., Casey G., Gallinger S., Le Marchand L., Lindor N.M., Newcomb P.A., Haile R.W., Colon Cancer Family Registry Cohort I Cohort profile: the colon cancer family registry cohort (CCFRC) Int J Epidemiol. 2018;47:387–388i. doi: 10.1093/ije/dyy006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Newcomb P.A., Baron J., Cotterchio M., Gallinger S., Grove J., Haile R., Hall D., Hopper J.L., Jass J., Le Marchand L., Limburg P., Lindor N., Potter J.D., Templeton A.S., Thibodeau S., Seminara D., Colon Cancer Family R. Colon Cancer Family Registry: an international resource for studies of the genetic epidemiology of colon cancer. Cancer Epidemiol Biomarkers Prev. 2007;16:2331–2343. doi: 10.1158/1055-9965.EPI-07-0648. [DOI] [PubMed] [Google Scholar]
- 44.Buchanan D.D., Clendenning M., Rosty C., Eriksen S.V., Walsh M.D., Walters R.J., Thibodeau S.N., Stewart J., Preston S., Win A.K., Flander L., Ouakrim D.A., Macrae F.A., Boussioutas A., Winship I.M., Giles G.G., Hopper J.L., Southey M.C., English D., Jenkins M.A. Tumour testing to identify Lynch syndrome in two Australian colorectal cancer cohorts. J Gastroenterol Hepatol. 2017;32:427–438. doi: 10.1111/jgh.13468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walsh M.D., Buchanan D.D., Pearson S.-A., Clendenning M., Jenkins M.A., Win A.K., Walters R.J., Spring K.J., Nagler B., Pavluk E., Arnold S.T., Goldblatt J., George J., Suthers G.K., Phillips K., Hopper J.L., Jass J.R., Baron J.A., Ahnen D.J., Thibodeau S.N., Lindor N., Parry S., Walker N.I., Rosty C., Young J.P. Immunohistochemical testing of conventional adenomas for loss of expression of mismatch repair proteins in Lynch syndrome mutation carriers: a case series from the Australasian site of the colon cancer family registry. Mod Pathol. 2012;25:722–730. doi: 10.1038/modpathol.2011.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cicek M.S., Lindor N.M., Gallinger S., Bapat B., Hopper J.L., Jenkins M.A., Young J., Buchanan D., Walsh M.D., Le Marchand L., Burnett T., Newcomb P.A., Grady W.M., Haile R.W., Casey G., Plummer S.J., Krumroy L.A., Baron J.A., Thibodeau S.N. Quality assessment and correlation of microsatellite instability and immunohistochemical markers among population- and clinic-based colorectal tumors results from the Colon Cancer Family Registry. J Mol Diagn. 2011;13:271–281. doi: 10.1016/j.jmoldx.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buchanan D.D., Tan Y.Y., Walsh M.D., Clendenning M., Metcalf A.M., Ferguson K., Arnold S.T., Thompson B.A., Lose F.A., Parsons M.T., Walters R.J., Pearson S.A., Cummings M., Oehler M.K., Blomfield P.B., Quinn M.A., Kirk J.A., Stewart C.J., Obermair A., Young J.P., Webb P.M., Spurdle A.B. Tumor mismatch repair immunohistochemistry and DNA MLH1 methylation testing of patients with endometrial cancer diagnosed at age younger than 60 years optimizes triage for population-level germline mismatch repair gene mutation testing. J Clin Oncol. 2014;32:90–100. doi: 10.1200/JCO.2013.51.2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weisenberger D.J., Siegmund K.D., Campan M., Young J., Long T.I., Faasse M.A., Kang G.H., Widschwendter M., Weener D., Buchanan D., Koh H., Simms L., Barker M., Leggett B., Levine J., Kim M., French A.J., Thibodeau S.N., Jass J., Haile R., Laird P.W. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 49.Zaidi S.H., Harrison T.A., Phipps A.I., Steinfelder R., Trinh Q.M., Qu C., et al. Landscape of somatic single nucleotide variants and indels in colorectal cancer and impact on survival. Nat Commun. 2020;11:3644. doi: 10.1038/s41467-020-17386-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Belhadj S., Terradas M., Munoz-Torres P.M., Aiza G., Navarro M., Capellá G., Valle L. Candidate genes for hereditary colorectal cancer: mutational screening and systematic review. Hum Mutat. 2020;41:1563–1576. doi: 10.1002/humu.24057. [DOI] [PubMed] [Google Scholar]
- 51.Seifert B.A., McGlaughon J.L., Jackson S.A., Ritter D.I., Roberts M.E., Schmidt R.J., Thompson B.A., Jimenez S., Trapp M., Lee K., Plon S.E., Offit K., Stadler Z.K., Zhang L., Greenblatt M.S., Ferber M.J. Determining the clinical validity of hereditary colorectal cancer and polyposis susceptibility genes using the Clinical Genome Resource Clinical Validity Framework. Genet Med. 2019;21:1507–1516. doi: 10.1038/s41436-018-0373-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weren R.D.A., Ligtenberg M.J.L., Kets C.M., de Voer R.M., Verwiel E.T.P., Spruijt L., van Zelst-Stams W.A.G., Jongmans M.C., Gilissen C., Hehir-Kwa J.Y., Hoischen A., Shendure J., Boyle E.A., Kamping E.J., Nagtegaal I.D., Tops B.B.J., Nagengast F.M., Geurts van Kessel A., van Krieken J.H.J.M., Kuiper R.P., Hoogerbrugge N. A germline homozygous mutation in the base-excision repair gene NTHL1 causes adenomatous polyposis and colorectal cancer. Nat Genet. 2015;47:668–671. doi: 10.1038/ng.3287. [DOI] [PubMed] [Google Scholar]
- 53.Spurdle A.B., Bowman M.A., Shamsani J., Kirk J. Endometrial cancer gene panels: clinical diagnostic vs research germline DNA testing. Mod Pathol. 2017;30:1048–1068. doi: 10.1038/modpathol.2017.20. [DOI] [PubMed] [Google Scholar]
- 54.Levine D.A. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cherniack A.D., Shen H., Walter V., Stewart C., Murray B.A., Bowlby R., Hu X., Ling S., Soslow R.A., Broaddus R.R., Zuna R.E., Robertson G., Laird P.W., Kucherlapati R., Mills G.B., Cancer Genome Atlas Research Network, Weinstein J.N., Zhang J., Akbani R., Levine D.A. Integrated molecular characterization of uterine carcinosarcoma. Cancer Cell. 2017;31:411–423. doi: 10.1016/j.ccell.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saunders C.T., Wong W.S.W., Swamy S., Becq J., Murray L.J., Cheetham R.K. Strelka: accurate somatic small-variant calling from sequenced tumor–normal sample pairs. Bioinformatics. 2012;28:1811–1817. doi: 10.1093/bioinformatics/bts271. [DOI] [PubMed] [Google Scholar]
- 58.Sha D., Jin Z., Budzcies J., Kluck K., Stenzinger A., Sinicrope F.A. Tumor mutational burden (TMB) as a predictive biomarker in solid tumors. Cancer Discov. 2020;10:1808–1825. doi: 10.1158/2159-8290.CD-20-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tibshirani R. Regression shrinkage and selection via the Lasso. J R Stat Soc Ser B Methodol. 1996;58:267–288. [Google Scholar]
- 60.Wickham H., Averick M., Bryan J., Chang W., McGowan L.D., François R., Grolemund G., Hayes A., Henry L., Hester J., Kuhn M., Pedersen T.L., Miller E., Bache S.M., Müller K., Ooms J., Robinson D., Seidel D.P., Spinu V., Takahashi K., Vaughan D., Wilke C., Woo K., Yutani H. Welcome to the tidyverse. J Open Source Softw. 2019;4:1686. [Google Scholar]
- 61.Robin X., Turck N., Hainard A., Tiberti N., Lisacek F., Sanchez J.-C., Müller M. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Friedman J.H., Hastie T., Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 63.Thiele C., Hirschfeld G. cutpointr: Improved estimation and validation of optimal cutpoints in R. J Stat Softw. 2021;98:1–27. [Google Scholar]
- 64.Wickham H. The split-apply-combine strategy for data analysis. J Stat Softw. 2011;40:1–29. [Google Scholar]
- 65.Wickham H. Springer-Verlag; New York, NY: 2016. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- 66.Clopper C.J., Pearson E.S. The use of confidence or fiducial limits illustrated in the case of the binomial. Biometrika. 1934;26:404–413. [Google Scholar]
- 67.Renault V., Tubacher E., How-Kit A. In: Computational Methods for Precision Oncology. Laganà A., editor. Springer International Publishing; Cham, Switzerland: 2022. Assessment of microsatellite instability from next-generation sequencing data; pp. 75–100. [Google Scholar]
- 68.Salipante S.J., Scroggins S.M., Hampel H.L., Turner E.H., Pritchard C.C. Microsatellite instability detection by next generation sequencing. Clin Chem. 2014;60:1192–1199. doi: 10.1373/clinchem.2014.223677. [DOI] [PubMed] [Google Scholar]
- 69.Ratovomanana T., Cohen R., Svrcek M., Renaud F., Cervera P., Siret A., Letourneur Q., Buhard O., Bourgoin P., Guillerm E., Dorard C., Nicolle R., Ayadi M., Touat M., Bielle F., Sanson M., Rouzic P.L., Buisine M.-P., Piessen G., Collura A., Fléjou J.-F., Reyniès Ade, Coulet F., Ghiringhelli F., André T., Jonchère V., Duval A. Performance of next-generation sequencing for the detection of microsatellite instability in colorectal cancer with deficient DNA mismatch repair. Gastroenterology. 2021;161:814–826.e7. doi: 10.1053/j.gastro.2021.05.007. [DOI] [PubMed] [Google Scholar]
- 70.Roufas C., Georgakopoulos-Soares I., Zaravinos A. Molecular correlates of immune cytolytic subgroups in colorectal cancer by integrated genomics analysis. NAR Cancer. 2021;3:zcab005. doi: 10.1093/narcan/zcab005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zaravinos A., Roufas C., Nagara M., de Lucas Moreno B., Oblovatskaya M., Efstathiades C., Dimopoulos C., Ayiomamitis G.D. Cytolytic activity correlates with the mutational burden and deregulated expression of immune checkpoints in colorectal cancer. J Exp Clin Cancer Res. 2019;38:364. doi: 10.1186/s13046-019-1372-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen W., Pearlman R., Hampel H., Pritchard C.C., Markow M., Arnold C., Knight D., Frankel W.L. MSH6 immunohistochemical heterogeneity in colorectal cancer: comparative sequencing from different tumor areas. Hum Pathol. 2020;96:104–111. doi: 10.1016/j.humpath.2019.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The cross-validated areas under the curves (AUCs) for the top 10 performing tumor features to predict the DNA mismatch repair status in the whole-exome sequenced (WES) colorectal cancer (CRC) data set. Receiving operator curves presenting the AUCs for the top 10 performing tumor features, MSMuTect, MANTIS, MSISensor, MSIseq, insertion/deletion (INDEL) count, tumor mutational burden (TMB; calculated as mutations per megabase), tumor mutational signature (TMS) small insertion/deletion (ID) 2 + ID7, TMS single-base substitution (SBS) 15, TMS SBS20, and TMS SBS54, as determined from the WES CRC analysis. The 95% CIs for each feature are shown in red highlighting.
Tumor distribution for each of the top 10 predicting features, MSMuTect, MANTIS, MSISensor, MSIseq, insertion/deletion (INDEL) count, tumor mutational burden (TMB; calculated as mutations per megabase), tumor mutational signature (TMS) small insertion/deletion (ID) 2 + ID7, TMS single-base substitution (SBS) 15, TMS SBS20, and TMS SBS54, identified from the whole-exome sequencing colorectal cancer (CRC) analysis that were applied to targeted panel-sequenced CRCs. The left of the vertical dotted line shows boxplots comparing the distribution of tumors by DNA mismatch repair (MMR) status: MMR proficient (pMMR) and MMR deficient (dMMR). The right of the vertical dotted line shows boxplots for each of the dMMR subgroups: dMMR–Lynch syndrome (LS), dMMR–double-somatic MMR gene mutation (DS), and dMMR–MLH1 promoter methylation (MLH1me).
Tumor distribution for each of the top 10 predicting features, MSMuTect, MANTIS, MSISensor, MSIseq, insertion/deletion (INDEL) count, tumor mutational burden (TMB; calculated as mutations per megabase), tumor mutational signature (TMS) small insertion/deletion (ID) 2 + ID7, TMS single-base substitution (SBS) 15, TMS SBS20, and TMS SBS54, identified from the whole-exome sequencing colorectal cancer analysis that were applied to targeted panel-sequenced endometrial cancers. The left of the vertical dotted line shows boxplots comparing the distribution of tumors by DNA mismatch repair (MMR) status: MMR proficient (pMMR) and MMR deficient (dMMR). The right of the vertical dotted line shows boxplots for each of the dMMR subgroups: dMMR–Lynch syndrome (LS), dMMR–double-somatic MMR gene mutation (DS), and dMMR–MLH1 promoter methylation (MLH1me).
Tumor distribution for each of the top 10 predicting features, MSMuTect, MANTIS, MSISensor, MSIseq, insertion/deletion (INDEL) count, tumor mutational burden (TMB; calculated as mutations per megabase), tumor mutational signature (TMS) small insertion/deletion (ID) 2 + ID7, TMS single-base substitution (SBS) 15, TMS SBS20, and TMS SBS54, identified from the whole-exome sequencing colorectal cancer analysis that were applied to targeted panel-sequenced sebaceous skin tumors. The left of the vertical dotted line shows boxplots comparing the distribution of tumors by DNA mismatch repair (MMR) status: MMR proficient (pMMR) and MMR deficient (dMMR). The right of the vertical dotted line shows boxplots for each of the dMMR subgroups: dMMR–Lynch syndrome (LS), dMMR–double-somatic MMR gene mutation (DS), and dMMR–MLH1 promoter methylation (MLH1me).
Determination of the thresholds for the top 10 performing features for panel colorectal cancers (CRCs). Bar graphs presenting the distribution of tumors after applying the recommended thresholds for each of the top 10 predicting tumor features, MSMuTect, MANTIS, MSISensor, MSIseq, insertion/deletion (INDEL) count, tumor mutational burden (TMB; calculated as mutations per megabase), tumor mutational signature (TMS) small insertion/deletion (ID) 2 + ID7, TMS single-base substitution (SBS) 15, TMS SBS20, and TMS SBS54, for targeted panel-sequenced CRCs. Orange indicates DNA mismatch repair (MMR) proficient (pMMR), and blue represents MMR-deficient (dMMR) status.
Determination of the thresholds for the top 10 performing features for panel endometrial cancers (ECs). Bar graphs presenting the distribution of tumors after applying the recommended thresholds for each of the top 10 predicting tumor features, MSMuTect, MANTIS, MSISensor, MSIseq, insertion/deletion (INDEL) count, tumor mutational burden (TMB; calculated as mutations per megabase), tumor mutational signature (TMS) small insertion/deletion (ID) 2 + ID7, TMS single-base substitution (SBS) 15, TMS SBS20, and TMS SBS54, for targeted panel-sequenced ECs. Orange indicates DNA mismatch repair (MMR) proficient (pMMR), and blue represents MMR-deficient (dMMR) status.
Determination of the thresholds for the top 10 performing features for panel sebaceous skin tumors (SSTs). Bar graphs presenting the distribution of tumors after applying the recommended thresholds for each of the top 10 predicting tumor features, MSMuTect, MANTIS, MSISensor, MSIseq, insertion/deletion (INDEL) count, tumor mutational burden (TMB; calculated as mutations per megabase), tumor mutational signature (TMS) small insertion/deletion (ID) 2 + ID7, TMS single-base substitution (SBS) 15, TMS SBS20, and TMS SBS54, for targeted panel-sequenced SSTs. Orange indicates DNA mismatch repair (MMR) proficient (pMMR), and blue represents MMR-deficient (dMMR) status.
The additive tumor feature combination approach demonstrating the distribution of counts of the top six tumor features by the DNA mismatch repair (MMR) status of the 29 panel-sequenced colorectal cancers (CRCs). Bar graphs presenting the distribution of tumors after applying the additive tumor feature combination approach with the recommended thresholds for targeted panel-sequenced CRCs using a count of three or more of the top six predictors with >95% accuracy, consisting of MSMuTect, MANTIS, MSIseq, MSISensor, insertion/deletion count, and tumor mutational signature small insertion/deletion (ID) 2 + ID7 for MMR status calling: MMR deficient (dMMR) versus MMR proficient (pMMR).
The additive tumor feature combination approach demonstrating the distribution of counts of the top six tumor features by the DNA mismatch repair (MMR) status of the 22 panel-sequenced endometrial cancers (ECs). Bar graphs presenting the distribution of tumors after applying the additive tumor feature combination approach with the recommended thresholds for targeted panel-sequenced ECs using a count of three or more of the top six predictors with >95% accuracy, consisting of MSMuTect, MANTIS, MSIseq, MSISensor, insertion/deletion count, and tumor mutational signature small insertion/deletion (ID) 2 + ID7 for MMR status calling: MMR deficient (dMMR) versus MMR proficient (pMMR).
The additive tumor feature combination approach demonstrating the distribution of counts of the top six tumor features by the DNA mismatch repair (MMR) status of the 20 panel-sequenced sebaceous skin tumors (SSTs). Bar graphs presenting the distribution of tumors after applying the additive tumor feature combination approach with the recommended thresholds for targeted panel-sequenced SSTs using a count of three or more of the top six predictors with >95% accuracy, consisting of MSMuTect, MANTIS, MSIseq, MSISensor, insertion/deletion count, and tumor mutational signature small insertion/deletion (ID) 2 + ID7 for MMR status calling: MMR deficient (dMMR) versus MMR proficient (pMMR).
Data Availability Statement
The data generated during and/or analyzed during the current study are included in this published article (and its supplemental information files/source data file). These data are available from the Colon Cancer Family Registry via a request to collaborate with the Colon Cancer Family Registry application process approved by the NIH/National Cancer Institute (http://www.coloncfr.org/collaboration, last accessed October 21, 2022).