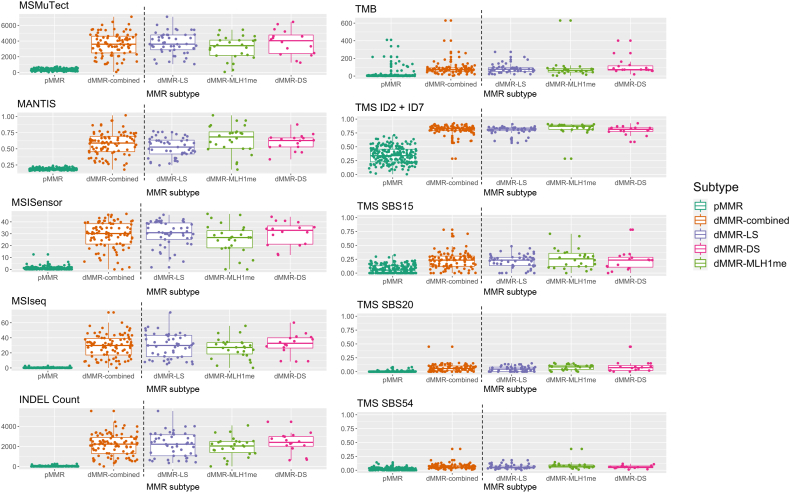
Figure 2.
Tumor distribution for each of the top 10 predicting features, MSMuTect, MANTIS, MSISensor, MSIseq, insertion/deletion (INDEL) count, tumor mutational burden (TMB; calculated as mutations/megabase), tumor mutational signature (TMS) small insertion/deletion (ID) 2 + ID7, TMS single-base substitution (SBS) 15, TMS SBS20, and TMS SBS54, as determined from the whole-exome sequencing analysis of colorectal cancers. The left of the vertical dotted line shows boxplots comparing the distribution of tumors by DNA mismatch repair (MMR) status: MMR proficient (pMMR) and MMR deficient (dMMR). The right of the vertical dotted line shows boxplots for each of the dMMR subgroups: dMMR–Lynch syndrome (LS), dMMR–double-somatic MMR gene mutation (DS), and dMMR–MLH1 promoter methylation (MLH1me).