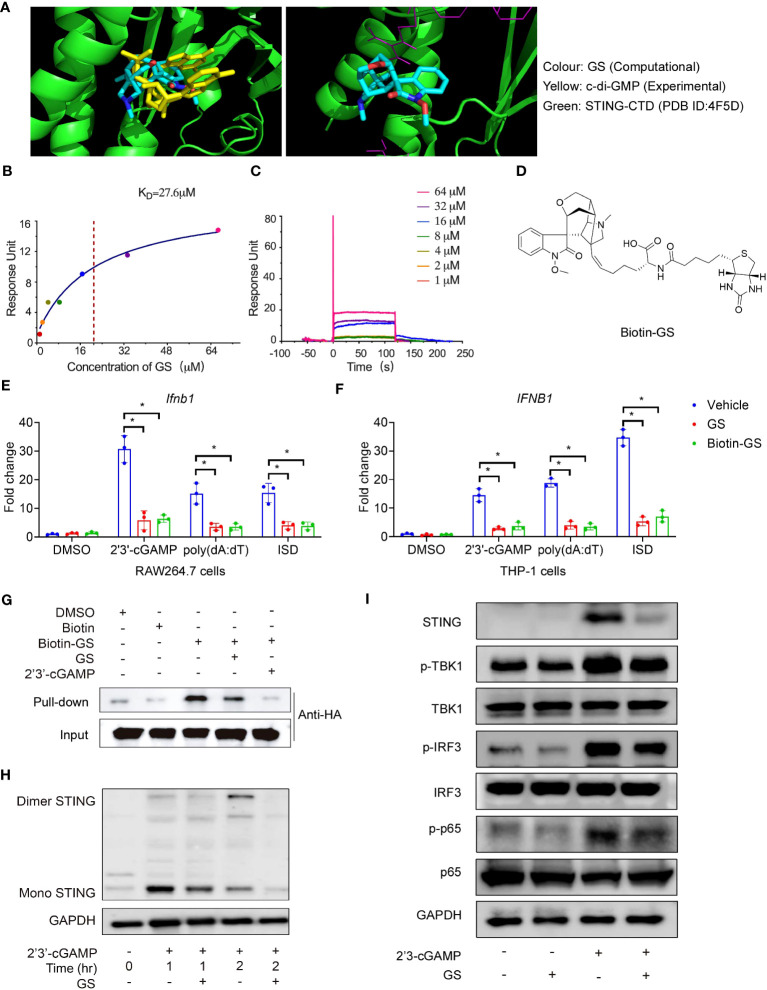
Figure 2.
Gelsevirine (GS) binds to the STING and inhibits STING activation. The in silico virtual simulation analysis of the binding between STING-CTD dimer (4F5D) and GS compared with c-di-GMP (A, left); the in silico simulation analysis of the binding site of the STING-CTD dimer (4F5D) with GS (A, right). Biacore analysis of hSTING-CTD and GS binding. Fitting the binding data to a steady-state 1:1 binding model yielded the binding affinity (Kd) (B, C). Chemical structures of biotin-GS (D). Raw264.7 and THP-1 cells were pretreated with GS (10μM) or biotin-GS (10μM) for 6 hrs and then stimulated with 2’3’-cGAMP (5 μg/ml), ISD (2 μg/ml), or Poly(dA:dT) (5 μg/ml) for 3 hrs. The mRNA expression of Ifnb1 (E) in Raw264.7 cells and mRNA expression of IFNB1 (F) in THP-1 cells were measured by RT-PCR. Cell lysates of HEK293T cells, transfected for 24 hrs with expression plasmids for the HA-tagged STING, were incubated with biotin (5 μM) or biotinylated GS (5 μM) for 1 hr with or without a 10-fold excess (50 μM) of GS or 2’3’-cGAMP (50 μg/ml), followed by pull-down with streptavidin-conjugated beads and immunoblot with anti-HA (G). Raw264.7 cells were pretreated with GS (10 μM) for 6 hrs and then stimulated with 2’3’-cGAMP for 1 hr or 2 hrs. STING dimerization (H) was analyzed by immunoblot. Raw264.7 was pretreated with GS (10 μM) for 6 hrs and then stimulated with 2’3’-cGAMP (5 μg/ml) for 3 hrs. The protein expression of STING and phosphorylation of TBK1, IRF3, and p65 was determined by Western blot (I). *P < 0.05 vs vehicle group.