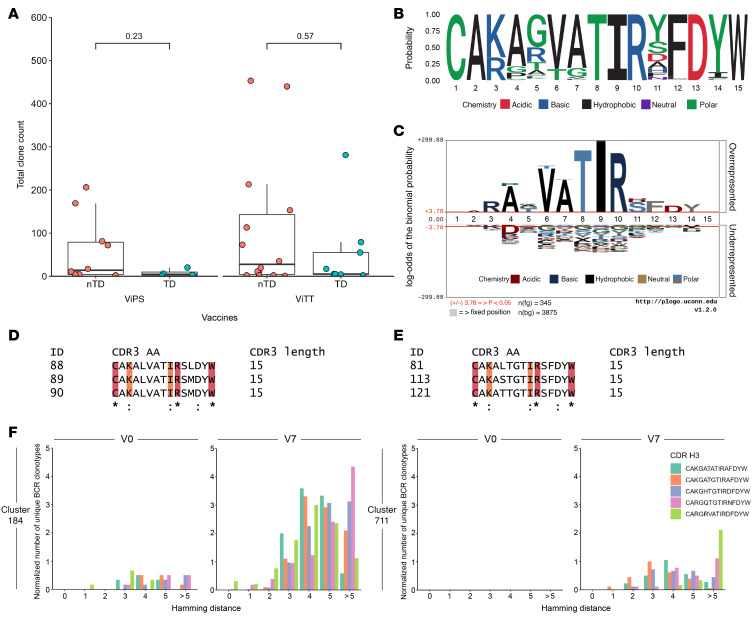
Figure 6. Amino acid sequence conservation of BCR cluster 184.
(A) Expression of cluster 184 in ViTT and ViPS recipients at V7. (B) Logo plot highlighting probability and amino acid residue properties. (C) pLogo plot demonstrating that most amino acid residues are significantly outstanding from the background. Significance was determined by Mann–Whitney U test; the red lines indicate a significance level cutoff of P < 0.05. (D and E) Amino acid sequence conservation calculated by Clustal Omega (19) between (D) cluster 184 CDR H3 sequences alone and (E) cluster 184 CDR H3 sequences and known Vi-binding BCR CDR H3 sequences. Three example sequences for each are included for simplicity. An asterisk denotes positions that have a single, fully conserved residue; a colon denotes amino acids with strong similarity, which scores higher than 0.5 in the Gonnet PAM 250 matrix. Conservation is highlighted in crimson (*) and orange (:). (F) Unique BCR clonotypes in cluster 184 and 711 and the relative proximity to known Vi-binding BCR CDR H3 sequences at V0 and V7 for ViPS and ViTT participants. The number of unique BCR clonotypes was normalized by the number of participants for each time point.