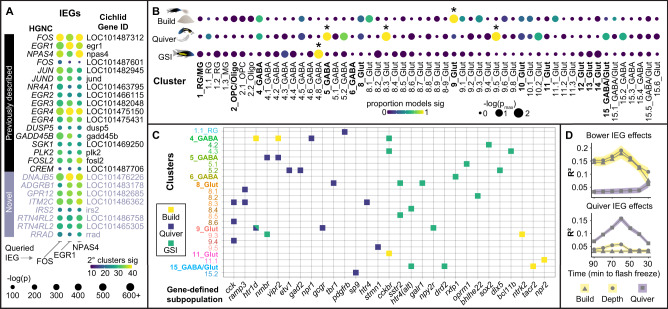
Fig. 3. Distinct cell populations exhibit building-, quivering-, and gonadal-associated IEG expression.
A Twenty-five genes were selectively co-expressed with c-fos, egr1, and npas4 across cell populations; genes that were detected in the majority of clusters, and that were significantly (p < 0.05, Wilcoxon rank-sum test, two-sided) upregulated in IEG-positive nuclei compared to IEG-negative nuclei in the majority of those clusters were considered significant. B Building-, quivering-, and gonadal-associated IEG expression was observed in distinct clusters (linear mixed-effects regression, two-sided, adjusted for 5% false discovery rate), and C gene-defined populations (filled squares indicate significant effects, linear mixed-effects regression, two-sided, adjusted for 5% false discovery rate; 9_Glut htr1d+ nuclei showed both quivering- and GSI-associated IEG expression). D IEG expression was most strongly associated with the amount of temporally binned building (top, 30 min bins, n = 22 neuronal populations exhibiting significant or trending building-associated IEG expression) and quivering (bottom, 30 min bins, n = 71 neuronal populations exhibiting significant or trending quiver-associated IEG expression) behavior performed approximately 60 min prior to tissue freezing (peak bins represent behavior performed 45–75 min prior to freezing the telencephalon). Data were presented as mean values ± SEM (represented by colored bands). Source data are provided as a Source Data file, and additional related data can be found in Supplementary Data 6. Fish artwork in panel B is reprinted from iScience, Vol 23 / Issue 10, Lijiang Long, Zachary V. Johnson, Junyu Li, Tucker J. Lancaster, Vineeth Aljapur, Jeffrey T. Streelman, Patrick T. McGrath, Automatic Classification of Cichlid Behaviors Using 3D Convolutional Residual Networks, 2020, with permission from Elsevier.