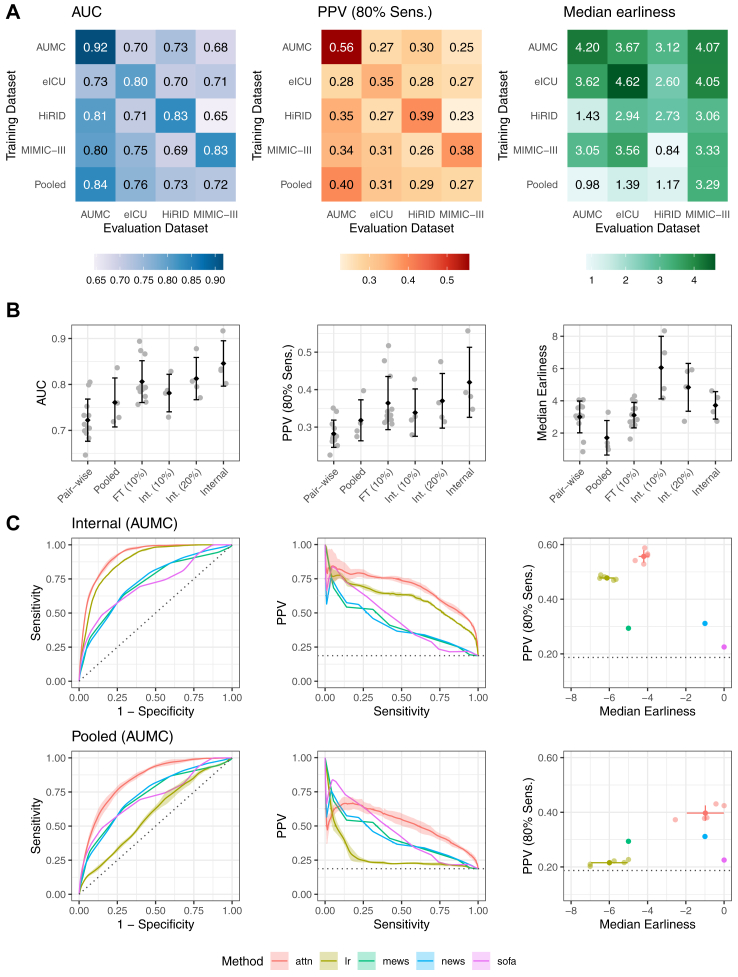
Fig. 3.
Performance of the deep learning system for the prediction of sepsis. In Panel A, heatmaps of the performance of our deep learning system are shown for AUC as well as PPV and median Earliness, whereas the latter two metrics are displayed at 80% Sensitivity. For a given heatmap, the rows indicate the training dataset, the columns refer to the testing dataset. In the last row, the externally pooled predictions are shown. Averaged across datasets, we observe an internally validated AUC of 0.846 (95% CI, 0.841–0.852), PPV of 42.0% (95% CI, 40.5–44.1), and median lead time to sepsis onset of 3.7 (95% CI, 3.0–4.3) hours. In the bottom row of the heatmaps, we observed an average externally validated AUC of 0.761 (95% CI, 0.746–0.770), PPV of 31.8% (95% CI, 30.3–34.0), and median lead time to sepsis onset of 1.71 (95% CI, 0.75–2.69) hours. The pooling approach improved the generalisability to new datasets by outperforming or being on par with predictions derived from the best (a priori unknown) training dataset in terms of AUC and PPV at 80% Sensitivity. Panel B illustrates how the metrics behave with different model transfer strategies: AUC performance of the internal validaton is increasingly approached when using pooling of models (pooled), and more so when instead fine-tuning a model to a small fine-tuning set of 10% of the testing site (FT (10%)), reflecting the realistic scenario that only small sample has been collected in a novel target hospital. By comparison, Int. (10%) and Int. (20%) displays the internal validation performance when training on only 10% or 20% of the training site, respectively. The error bars indicate the standard deviation of the metrics as calculated over the four datasets. A black diamond indicates the mean over the datasets. In Panel C, more detailed performance curves of the internal validation (top row) and external validation via pooled predictions (bottom row) performance are shown for an example dataset (AUMC). Our deep learning approach (attn) is visualised together with a subset of all included baselines, including clinical baselines (SOFA, MEWS, NEWS), and LASSO-regularised logistic regression (lr). Error bands indicate standard deviation over 5 repetitions of train-validation splitting. All baselines are shown in Supplementary Figs. S4–S7.