Comprehensive characterization of the five subtypes of metastatic castration-resistant prostate cancer identified transcription factors that drive each subtype and showed that the double-negative subtype has the worst prognosis.
Abstract
Systemic targeted therapy in prostate cancer is primarily focused on ablating androgen signaling. Androgen deprivation therapy and second-generation androgen receptor (AR)–targeted therapy selectively favor the development of treatment-resistant subtypes of metastatic castration-resistant prostate cancer (mCRPC), defined by AR and neuroendocrine (NE) markers. Molecular drivers of double-negative (AR−/NE−) mCRPC are poorly defined. In this study, we comprehensively characterized treatment-emergent mCRPC by integrating matched RNA sequencing, whole-genome sequencing, and whole-genome bisulfite sequencing from 210 tumors. AR−/NE− tumors were clinically and molecularly distinct from other mCRPC subtypes, with the shortest survival, amplification of the chromatin remodeler CHD7, and PTEN loss. Methylation changes in CHD7 candidate enhancers were linked to elevated CHD7 expression in AR−/NE+ tumors. Genome-wide methylation analysis nominated Krüppel-like factor 5 (KLF5) as a driver of the AR−/NE− phenotype, and KLF5 activity was linked to RB1 loss. These observations reveal the aggressiveness of AR−/NE− mCRPC and could facilitate the identification of therapeutic targets in this highly aggressive disease.
Significance:
Comprehensive characterization of the five subtypes of metastatic castration-resistant prostate cancer identified transcription factors that drive each subtype and showed that the double-negative subtype has the worst prognosis.
Introduction
Although localized prostate cancer is usually well controlled by radiation, surgery, or systemic androgen deprivation therapy (ADT), metastatic prostate cancer has a 5-year survival rate of only 31% (1). Hormone-refractory metastatic disease, known as castration-resistant prostate cancer (CRPC), develops after tumors become resistant to ADT (2). Progression to metastatic CRPC (mCRPC) is associated with recurrent driver gene alterations. In approximately 80% of cases, somatic alterations affect the androgen receptor (AR) itself or a nearby AR enhancer locus (3–5). Many patients with mCRPC receive AR-targeting therapies such as enzalutamide or abiraterone acetate. Progression on these therapies is associated with further AR alterations (6). However, a subset of treatment-resistant mCRPC infrequently harbors AR somatic alterations and instead develops lineage features of small cell neuroendocrine carcinoma (7–12). Patients whose tumors have this phenotype have worse prognosis than those with adenocarcinoma mCRPC (8). It was recently proposed that five distinctive histologic and expression-based subtypes of mCRPC exist (13): adenocarcinoma (AR+/NE−), double-positive (AR+/NE+), low AR (ARL/NE−), neuroendocrine (AR−/NE+), and double-negative (AR−/NE−). While these subtypes have been described at the transcriptional level, the etiology and clinical implications of the low AR and double-negative subtypes are largely unknown. Herein, we define the somatic alterations and DNA-methylation changes among these five subtypes by integrating whole transcriptome RNA sequencing (RNA-seq), whole-genome sequencing (WGS), and whole-genome bisulfite sequencing (WGBS) from 210 mCRPC tumors.
Materials and Methods
Tumor specimens
Image-guided fresh-frozen mCRPC biopsy acquisition and DNA extraction were performed as previously described (5, 11). WGS and WGBS libraries were prepared and processed as previously described (5, 11). The clinical characteristics of patients in this study are available in Supplementary Table S1. Human studies were approved and overseen by the UCSF Institutional Review Board (IRB) in accordance with the Declaration of Helsinki. All individuals provided written informed consent to obtain fresh tumor biopsies and to perform comprehensive molecular profiling of tumor and germline samples.
Data processing
RNA-seq data derived from laser-capture micro-dissected samples were aligned with STAR (14). RNA abundance was calculated using the default parameters, and transcripts were quantified at the gene level by GENECODE v.28, as previously described (11). The expression level of each gene was then converted to transcripts per million (TPM). WGBS data were aligned to GRCh38, and de-duplication, then base-level methylation calling was performed using Bismark 0.23.0 with “–pairedend” and “–no_overlap” parameters set; otherwise, default parameters were used, as recommended by the Bismark User Guide for the library kit.
Statistical analysis
All statistical analyses were conducted using the R statistical software version 4.2.0. Hierarchical clustering was performed using Ward's linkage algorithm with Euclidean distances. Survival analysis was performed using the survival package in R and survival probability was visualized using the Kaplan–Meier method, with endpoint overall survival defined from the time biopsies were obtained from the patients to death from any cause. All correlation analyses were performed using Pearson's method unless otherwise specified. Fisher exact test was applied to determine if DNA alterations were significantly different between the subtype groups. All tests were 2-sided when applicable, and P < 0.05 was considered statistically significant. Results were corrected for multiple testing using the Benjamini-Hochberg method (FDR) unless otherwise stated. All measurements were taken from distinct individual samples. Box plots should be interpreted as follows: horizontal lines denote median values; boxes extend from the 25th to the 75th percentile of each group's distribution of values; vertical extending lines denote adjacent values (the most extreme values within 1.5 interquartile range of the 25th and 75th percentile of each group). Differences between groups were assessed by the Kruskal–Wallis test. Significance is indicated as follows in the figures: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
Differentially expressed gene analyses
Differential gene expression analysis was performed using RNA-seq raw feature counts with DESeq2 version 1.36.0 (15). The data were corrected for tumor purity and tumor ploidy. Genes with fold change ≥ 2 or ≤ −2 and FDR ≤ 0.01 were considered significantly up- or downregulated, respectively.
Evaluation of copy-number alteration and tumor purity and ploidy
The PURPLE tool (16) was used on WGS data to evaluate copy-number alterations and assess the tumor purity and tumor ploidy. Copy number and biallelic status of the tumors were determined by incorporating tumor purity, tumor ploidy, and chromosome type (autosomal or sex chromosome). Genes were classified as amplified or deleted according to the following criteria: for the genes in chromosomes X and Y, a gene was marked as amplified if a minimum coding copy number was higher than tumor ploidy * 0.9. A gene was marked as a single copy deletion if the coding copy number was lower than 0.75. A gene was marked as two copies deleted if the maximum coding copy number was lower than 0.5. For genes in autosomal chromosomes, a gene was marked as amplified if a minimum coding copy number exceeded tumor ploidy * 1.95. Genes were marked as deleted if their minimum coding copy number was lower than 1.1. Genes were marked as two copies deleted if their maximum coding copy number was lower than 0.5. Copy-number bounds used in this analysis were determined by reviewing genome-wide distributions of all corrected gene copy estimates.
Evaluation of structural variants and mutation calling
Somatic mutation analysis was performed with Strelka2 version 2.9.10 and MuTect version 1.1.7 (17). Alterations with a PASS score in both tools were used to improve the accuracy of the results as recommended (18). SnpEff version 4.3 was used to identify Frameshift, Missense, Splice donor, Splice acceptor, Stop gain or Stop loss. Germline mutation analysis was performed using HaplotypeCaller version 4.2.2.0. GRIDSS version 2.12.2 and LINX (19) version 1.17 were used to identify structural variations and gene fusions, respectively. Samples lacking a PASS designation were excluded from the analyses.
Differentially methylated regions
Differential methylation analysis was performed using the DSS tool, version 2.26.0107 (20). No minimum CpG read coverage was set because DSS considers the read depth for calculating the differentially methylated regions (DMR). The smoothing was set to TRUE, otherwise, default parameters were used in DSS. DMRs were required to pass the following criteria: hypermethylated regions should have at least 10% higher methylation level and hypomethylated regions should have at least 10% lower methylation level in each subtype compared with the same regions in AR+/NE−. The same criteria were used to identify DMRs in AR+/NE− when compared with all other subtypes combined.
Motif analysis in DMR regions
A list of all known Homo sapiens transcription factor (TF) motifs was downloaded from the JASPAR database (21). This list was employed to perform an unbiased motif analysis using FIMO version 5.1.0 (22) with default parameters. FIMO was used to identify the occurrence of known motifs with potential regulatory functions that may bind the putative enhancer regions identified in the CHD7 gene. Regions of interest in the CHD7 gene (DMR2 and DMR3) on build GRCh38 were used as inputs in FIMO. Results were ranked by FDR (q value). DMRs including hyper- and hypomethylated regions identified by DSS for each subtype were converted to bed files using the GenomicRanges package version 1.48.0. We excluded ENCODE Blacklist (23) regions annotated in GRCh38, under accession number ENCFF419RSJ, and genomic coordinates outside of chromosomes 1–22, X, and Y. The BED files were used as inputs for the motif enrichment analyses using the HOMER program suite version 4.11.1 (24) (findMotifsGenome.pl) with “-size given”, otherwise default parameters. Significantly enriched motifs, were ranked by log (P value). The top 20 motifs, if available, within each subtype were plotted on heat maps. Genes mapped to Krüppel-like factor 5 (KLF5) were annotated using HOMER (annotatePeaks.pl).
Gene set enrichment analysis
We obtained gene sets of the Cancer Hallmark pathways from the Molecular Signatures Database (MSigDB) using msigdbr version 7.5.1 to conduct gene set enrichment analysis (GSEA) and single sample GSEA (ssGSEA). ssGSEA was carried out using GSVA version 1.44.1 (25). A matrix of RNA-seq read counts was used as an input and the recommended parameters were applied for the ssGSEA analysis (tau = 0.25, kcdf = "Poisson", method = "ssgsea"). In Gene Ontology (GO) enrichment analyses, differentially expressed genes unique to each subtype were ranked by their log2 (fold change) value, and the GO enrichment analyses were computed using the clusterProfiler R package version 4.4.2 (26) with default parameters. The gene sets with enrichment of FDR < 0.1 were considered significant. Genes annotated to the KLF5 TF using HOMER were ranked by FDR and GSEA was performed using the enrichR (27) tool with default parameters. The P values of enriched pathways were then adjusted for multiple testing using FDR. Pathways enriched with FDR < 0.1 were considered to be significant.
Code availability
Code used in this manuscript is available at https://github.com/DavidQuigley/WCDT_subtypes.
Data availability
RNA-seq FASTQ files of 148 localized samples from the CPC-GENE cohort (28) were obtained from the European Genome-Phenome Archive (EGA) under accession number EGAS00001000900 and the FASTQ files of eight benign samples from the PAIR cohort (29) were retrieved from Gene Expression Omnibus (GEO) database under accession number GSE115414. The files were aligned with STAR, and the gene level quantification was performed using gene models in GENECODE version 28. The expression value of each gene was converted to TPM. The chromatin immunoprecipitation sequencing (ChIP-seq) data of DNase I hypersensitive sites (DHS; ref. 30) was obtained from the ENCODE project under accession number ENCSR857UZV. The H3K27ac ChIP-seq data of primary prostate tumors (31) was obtained from GEO, under accession number GSE120738. WGBS and WGS from 100 samples of mCRPC tumors from the West Coast Prostate Cancer Dream Team (WCDT) cohort are available on dbGaP with study number phs001648 (11) and an additional 28 samples are available on EGA with study number EGAS00001006649. RNA-seq data from 210 samples of mCRPC tumors from the WCDT cohort are available on EGA with study numbers EGAD00001008991, EGAD00001008487, and EGAD00001009065 (Supplementary Table S2). All other raw data are available upon request from the corresponding author.
Results
Subtypes of mCRPC are associated with distinct transcriptional phenotypes
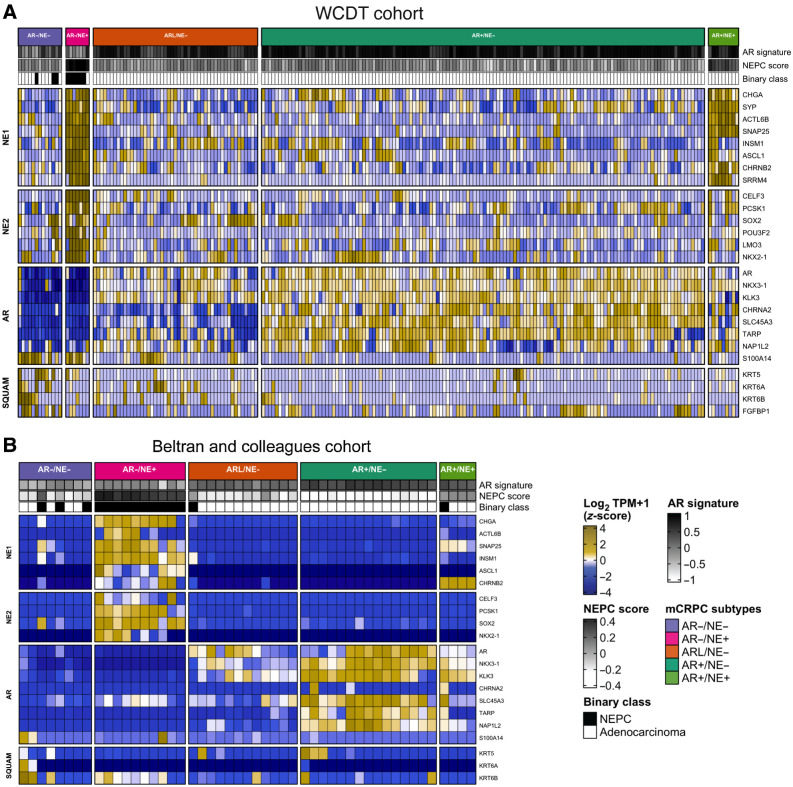
We developed a cohort of 210 mCRPC tumors from fresh-frozen core biopsies obtained through a prospective multi-institutional IRB-approved study (NCT02432001; ref. 8). All 210 tumors of the WCDT cohort were characterized by RNA-seq, with 128 tumors also characterized by WGS and WGBS. The clinical characteristics of patients in the cohort are listed in Supplementary Table S1, and characteristics of the molecular analysis are summarized in Supplementary Table S2. All samples were processed by a uniform analysis pipeline to evaluate transcriptional activity, somatic alterations, and tumor methylation status (Materials and Methods). We first tested the hypothesis that the AR and NE tumor subtypes identified in Labrecque and colleagues (13) could be replicated in this independent cohort. To this end, we clustered the WCDT gene expression data by employing a gene set previously demonstrated to distinguish these subtypes (13). Using hierarchical clustering we identified 132 tumors as AR+/NE−, 9 as AR+/NE+, 49 as ARL/NE−, 7 as AR−/NE+, and 13 as AR−/NE− (Fig. 1A). An unbiased genome-wide principal component analysis performed on tumor gene expression data identified clusters consistent with the supervised gene set clustering analysis (Supplementary Fig. S1). We inferred that the hierarchical clustering approach identified subtypes in the WCDT cohort consistent with those previously described by Labrecque and colleagues (13), and that these subtypes were associated with a large proportion of the overall transcriptional variance in our cohort. We repeated this analysis in an independent cohort of mCRPC tumors (7) and identified the same set of five transcriptionally defined subtypes (Fig. 1B), further supporting the generality of this subtype classification.
Figure 1.
mCRPC tumors cluster into five groups using the expression of androgen, neuroendocrine, and squamous gene panels. A and B, Heat map representing RNA-seq gene expression level of AR, NE, and squamous (SQUAM) gene panels of mCRPC tumors from the WCDT cohort (A; refs. 5, 11) and the Beltran and colleagues cohort (B; ref. 7). Results are expressed as log2 TPM (z-score) and colored from low (blue) to high (yellow) expression level. AR gene panel includes AR and AR-regulated genes, NE gene panels (NE1 and NE2) include NE related genes, and SQUAM panel includes genes associated with squamous cell differentiation. The expression levels of genes included in neuroendocrine prostate cancer (NEPC) panel from Beltran and colleagues cohort (7) were used to assign a binary classification (Binary class) of the samples based on their gene expression. White, adenocarcinoma tumors; black, small cell NEPC. AR and NEPC signature scores were calculated based on the AR and NEPC-related gene expression values as reported previously (7). The tumor subtypes can be read as follows: AR+/NE−, dark turquoise; ARL/NE−, dark orange; AR−/NE−, light purple; AR−/NE+, pink; AR+/NE+, light green.
We next asked whether these subtypes are present in localized tumors, or if they instead are exclusively observed in tumors that have progressed on ADT. We clustered gene expression data from eight benign samples from the PAIR cohort (29) and 148 localized prostate cancer samples from the CPC-GENE cohort (28) in addition to the WCDT mCRPC tumors using the Labrecque gene sets (13). Localized tumors were not associated with subtypes in this analysis (Supplementary Fig. S2). Six localized tumors with high levels of chromogranin-A (CHGA) expression, a neuroendocrine lineage marker, and low AR expression clustered with the mCRPC tumors, closer to NE+ and AR-low biopsies. This analysis was consistent with a model wherein these subtypes either arise de novo after progression on ADT or arise from rare cell populations among localized tumors that cannot be readily identified by bulk sequencing (32).
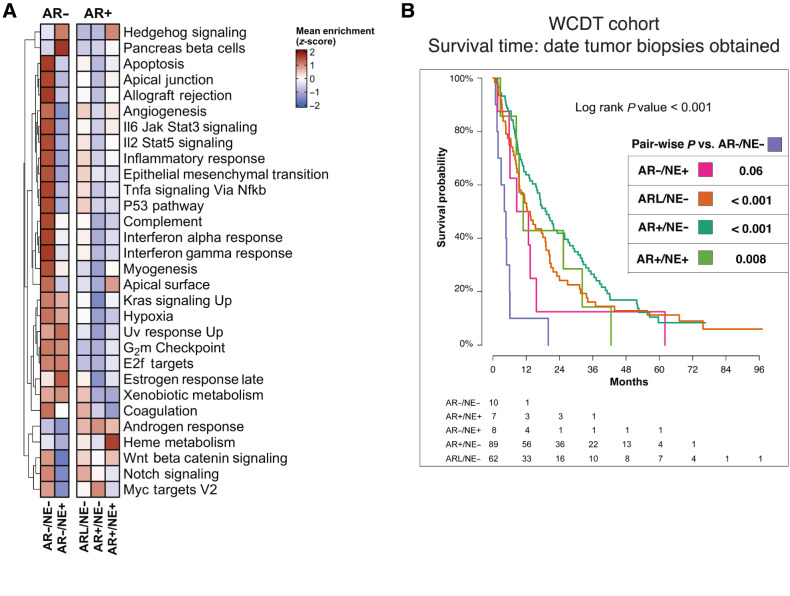
We next set out to identify the expression pathways that distinguish the subtypes. Consistent with previous studies, AR expression status was the major determinant in mCRPC molecular measurements (5, 9, 11) and was associated with the largest number of differentially expressed genes. In comparison with AR+/NE− tumors, we identified 1,557 and 2,856 differentially expressed genes specific to AR−/NE− and AR−/NE+ subtypes, respectively (Supplementary Fig. S3; Supplementary Data 1). AR+ subtypes were significantly enriched for androgen response, while NE+ subtypes were enriched for neuronal lineage and proliferation gene sets such as Hallmarks of Pancreas Beta Cells and E2F targets (Student t test P < 0.001; Fig. 2A; Supplementary Fig. S4). Tumors in AR− subtypes were enriched for hallmarks of hypoxia and proliferation (Student t test P < 0.001; Fig. 2A). Double-negative AR−/NE− tumors had downregulation of adaptive immune response genes, consistent with reports that this subtype has an immunosuppressed tumor microenvironment (33), and elevated expression of genes related to innate immune response and fibroblast growth factor signaling, as previously reported (Supplementary Fig. S4; ref. 34). Taken together, these data validate the presence of these mCRPC transcriptional subtypes in metastatic prostate tumors and demonstrate that these subtypes can be identified at a time when this knowledge could potentially lead to a change in therapy.
Figure 2.
Distinct clinical outcomes associated with the five subtypes of mCRPC. A, Heat map representing results of ssGSEAs and colored according to the legends. B, Kaplan–Meier curves representing clinical outcome of patients in the WCDT cohort, using survival from date of biopsy acquisition as the clinical outcome. Pairwise test conducted between AR−/NE− and other subtypes. The tumor subtypes can be read as follows: AR+/NE−, dark turquoise; ARL/NE−, dark orange; AR−/NE−, light purple; AR−/NE+, pink; AR+/NE+, light green.
The AR−/NE− subtype is associated with the worst prognosis
Neuroendocrine mCRPC, which has also been termed aggressive variant disease, is associated with poor patient outcomes (8, 35). We assessed the patient outcomes of the five molecular subtypes of mCRPC that we identified in the WCDT cohort of men with mCRPC. We tested for association between molecular subtypes and patients’ survival from the date tumor biopsies were obtained. Survival analyses confirmed that patients with AR− tumors had inferior overall outcomes relative to AR+ tumors (log-rank P < 0.001). There was not a significant association between AR signaling inhibitor exposure and either AR− status or individual tumor subtype (Supplementary Table S1). Notably, pairwise-comparisons tests between the AR− and AR+ subtypes indicated that the strongest significant difference in survival was associated with the AR−/NE− subtype (vs. AR+/NE− P < 0.001, vs. AR+/NE+ P = 0.008, vs. AR−/NE+ P = 0.06, vs. ARL/NE− P < 0.001; Fig. 2B).
Biallelic loss of PTEN is associated with the AR−/NE− subtype
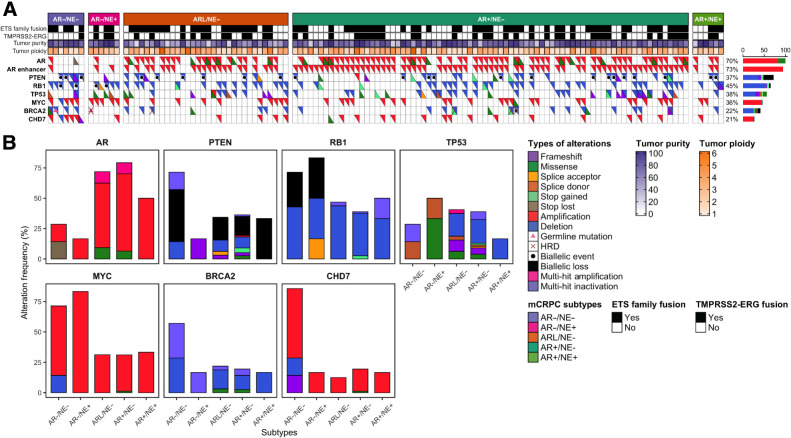
Leveraging the integrated molecular data available for the WCDT cohort, we tested for association between somatic alterations and the five mCRPC subtypes. We focused on 131 frequently altered prostate cancer driver genes (5, 36), and conducted somatic mutation and structural variation analyses to identify variants linked to each subtype. As expected, AR+ tumors harbored more frequent amplification of AR and a nearby AR enhancer than AR− tumors (AR amplified in 69% vs. 15% Fisher exact test P = 0.001 and AR enhancer amplified in 79% vs. 23%, Fisher exact test P < 0.001; Fig. 3A). Inactivation of the tumor suppressor genes TP53 and RB1 has been reported to be frequent in neuroendocrine prostate cancer (37). Combined biallelic loss of RB1 and TP53 alterations was significantly more frequent in AR− tumors than other subtypes (23% vs. 0%, Fisher exact test P = 0.002; Fig. 3A).
Figure 3.
Somatic and structural alterations associated with subtypes of mCRPC. A, Top rows show mCRPC subtypes, ETS family fusions, TMPRSS2-ERG fusions, tumor purity, and tumor ploidy in the WCDT cohort. Bottom rows show occurrence of AR, AR enhancer, PTEN, RB1, TP53, MYC, BRCA2, and CHD7 alterations in each sample. Tumors are sorted by their subtypes. Alteration frequency is shown to the right. B, Bar plots representing alteration frequency (%) of AR, PTEN, RB1, TP53, MYC, BRCA2, and CHD7 genes within each subtype. In both panels, types of alterations are colored (and/or marked with symbols) according to the legends.
Loss of the tumor suppressor gene PTEN has been associated with castration resistance and worse survival outcomes in response to AR-targeted therapy (38–40). We observed more frequent PTEN biallelic loss and inactivation in AR−/NE− tumors compared with the other subtypes (57%, AR−/NE− vs. 17%, Fisher exact test P = 0.031; Fig. 3A and B). Germline alterations inactivating an allele of BRCA2 are associated with more aggressive prostate cancer (41), and biallelic inactivation of homologous recombination repair genes including BRCA2 is predictive of response to PARP inhibitor therapy (42, 43). Two of the eight tumors with biallelic inactivation of BRCA2 were AR−/NE− (29% of AR−/NE− vs. 5% in other subtypes, Fisher exact test P = 0.061). MYC activation is a key driver of aggressive prostate cancer tumors and is associated with poor prognosis (44), and it has been observed that MYC overexpression impacts the activity of AR targets (45). We observed positive correlation between MYC copy gain and MYC gene expression level among the tumors (R = 0.3, P < 0.001). AR− tumors were more likely to harbor copy gain of MYC than AR+ tumors (69% in AR− vs. 29% in AR+, Fisher exact test P = 0.019; Fig. 3A and B). Gene fusions in the ETS family are the most common alterations in localized prostate cancer. 62% of the WCDT tumors harbored ETS fusions and was not associated with tumor subtypes (Supplementary Table S3). These results demonstrated that PTEN biallelic loss, previously associated with poor prognosis, was most frequently observed in AR−/NE− tumors compared with the other subtypes. These associations were consistent with our observation that AR−/NE− tumors were associated with the worst prognosis for WCDT patients (Fig. 2B).
Alterations in the chromatin remodeling gene CHD7 are associated with AR− tumors
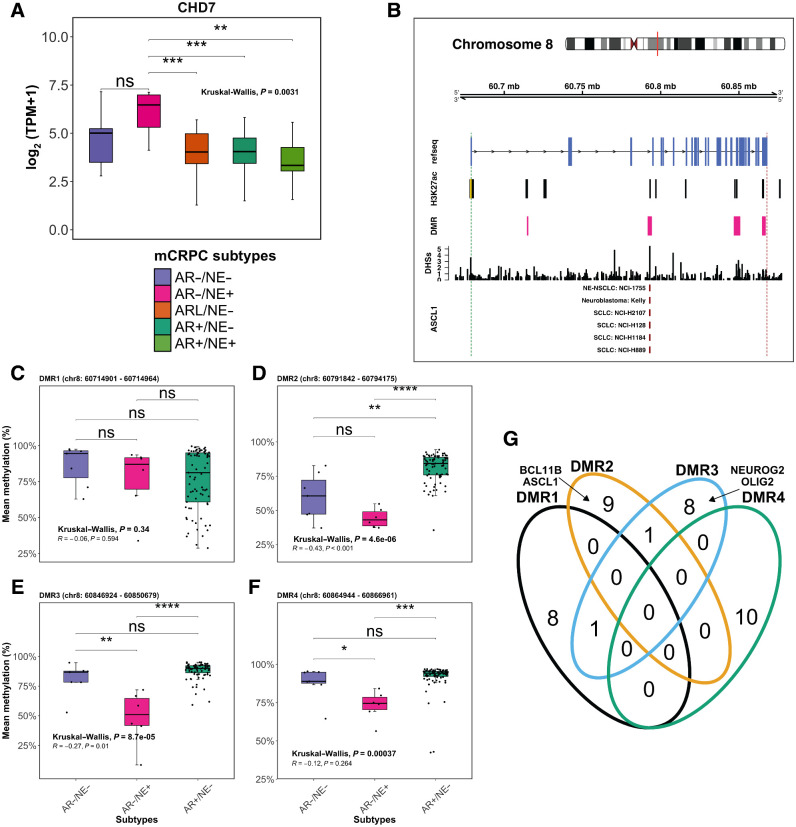
Of the 131 prostate cancer genes we examined, chromodomain helicase DNA binding protein 7 (CHD7) was the only gene with significantly higher copy numbers in the AR−/NE− tumors compared with the other subtypes (57% vs. 17%, Fisher exact test P = 0.031; Fig. 3A and B). Copy-number gain of CHD7, located at 8q12, was distinct from gain of MYC, located at 8q24. Notably, CHD7 expression was significantly higher in AR− tumors compared with AR+ tumors (Kruskal–Wallis, P = 0.0031; Fig. 4A) and was positively correlated with SOX2 expression (R = 0.25; P < 0.001; Supplementary Fig. S5). CHD7 was expressed at the highest levels in AR−/NE+ tumors, despite a very low rate of somatic alterations in this subtype (Fig. 3A and B). CHD7 was also expressed at significantly higher levels in AR−/NE+ tumors than other subtypes in an independent cohort (7) of mCRPC tumors (Supplementary Fig. S6). CHD7 is an ATP-dependent chromatin remodeler essential for multipotent neural crest formation (46). CHD7 plays a key role in promoting neural progenitor differentiation in embryonic stem cells (ESC), where it co-localizes with active gene enhancers such as SOX2 and subsequently modulates the expression of ESC-related genes (47–49). SOX2 plays an important role in disease progression, promoting androgen independence and lineage plasticity in prostate cancer (50–52). The consistent elevated expression of CHD7 in AR− tumors led us to hypothesize that CHD7 plays a role in AR− mCRPC.
Figure 4.
Hypomethylation in the putative enhancer regions of CHD7 is correlated with elevated gene expression in AR−/NE+. Integration of gene expression and DNA-methylation data for the CHD7 gene. A, Box plots representing CHD7 gene expression in the five mCRPC subtypes, colored according to the key below the plot. B, Top, chromosomal location of the CHD7 gene along with H3K27ac ChIP-seq marker, DHS, and DMRs in AR−/NE+ tumors compared with AR+/NE−. Bottom, ChIP-seq data for ASCL1 in different cell lines as indicated in the panels. The vertical dashed green and red lines show the transcription start site and transcription end site of the CHD7 gene, respectively. The yellow bar indicates the canonical promoter region of CHD7. C–F, Box plots showing mean methylation level per sample in DMR1 (C), DMR2 (D), DMR3 (E), and DMR4 (F) for AR−/NE−, AR−/NE+, and AR+/NE− subtypes. Pearson correlations were calculated between CHD7 gene expression and mean methylation of each sample at DMRs1–4. Box plots should be interpreted as follows: horizontal lines, median values; boxes extend from the 25th to the 75th percentile of each group's distribution of values; vertical extending lines, adjacent values (the most extreme values within 1.5 interquartile range of the 25th and 75th percentile of each group). Differences between groups were assessed by the Kruskal–Wallis test. Significance is indicated as follows: ns, not significant; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001. G, Venn diagram representing the overlap between the top 10 TF motifs enriched at each DMR location. Neuroendocrine-lineage motifs found in DMRs are labeled in the panel.
We observed that elevated CHD7 expression in AR−/NE+ tumors was not associated with increased CDH7 copy number; thus, we investigated the hypothesis that DNA methylation changes impact CHD7 expression in this subtype. DNA methylation plays a prominent role in the modulation of cellular states such as cell differentiation and tumorigeneses (53, 54). Increased methylation at DNA enhancer regions can reduce the expression of the targets of that enhancer (55, 56) by preventing TF binding (57–59). We tested for differential methylation at the CHD7 promoter and nearby genomic loci and predicted the presence of enhancers by intersecting these loci with regions marked by H3K27ac ChIP-seq in localized prostate tumors (31), and by DNase I sensitivity, assays that predict enhancer activity (Fig. 4B; ref. 30). We identified four statistically significant DMRs overlapping with H3K27ac ChIP-seq and DHS peaks. The loci were designated DMR1 (Chr8: 60714901–60714964), DMR2 (Chr8: 60791842–60794175), DMR3 (Chr8: 60846924–60850679), and DMR4 (Chr8: 60864944–60866961). DMR2 and DMR3 had 43% lower methylation levels in AR−/NE+ tumors compared with AR+/NE− tumors (Fig. 4C–F). Methylation levels in DMR2 and DMR3 were negatively correlated with CHD7 gene expression level, consistent with a role as enhancers of CHD7 expression (R = −0.43; P < 0.001 and R = −0.27; P = 0.010, respectively; Fig. 4D and E).
Having identified two candidate enhancer regions that are preferentially hypomethylated in AR−/NE+ compared with AR+/NE−, we next performed a DNA motif enrichment analysis on the DMR2 and DMR3 regions to identify TFs that may affect CHD7 expression. Unbiased motif enrichment analyses indicated that DMR2 was most significantly enriched for neuronal lineage TFs including BCL11B (q value = 0.003; ref. 60) and ASCL1 (q value = 0.009; ref. 10). In DMR3, NEUROG2 (q value = 0.006) and OLIG2 (q value = 0.01; refs. 10, 61) were the most significantly enriched TFs. In contrast, DMR1 and DMR4, whose methylation levels were not significantly correlated with CHD7 expression, do not contain these motifs (Fig. 4G). These data are consistent with a model in which hypomethylation at these neuroendocrine TF binding regions of CHD7 could contribute to the upregulation of CHD7 expression in AR−/NE+ tumors via binding of neuronal TFs such as ASCL1.
Expression and methylation analysis converges on KLF5 in AR−/NE− tumors
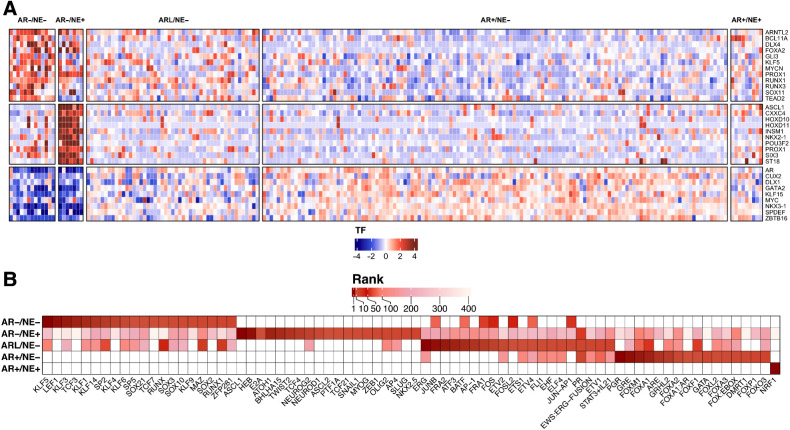
We next extended this analysis to nominate TFs that influence the development and activity in all mCRPC subtypes. We combined two orthogonal unbiased methods to identify the strongest candidates: subtype-specific differential expression analysis, and motif enrichment analysis at regions preferentially hypomethylated in each subtype. We hypothesized that subtype-specific driver TFs would be both upregulated and would have an increased number of hypomethylated binding sites in that subtype. Differential expression analysis across all subtypes, restricted to established TFs (62), identified subtype-specific upregulation of numerous TFs previously associated with AR+/NE− and AR−/NE− disease. As expected, AR+/NE− tumors expressed AR, GATA2, NKX3–1, and MYC at significantly higher levels than other subtypes (Fig. 5A). Consistent with prior reports, AR−/NE+ tumors had significantly higher expression of ASCL1, INSM1, and NKX2–1 (Fig. 5A). We then focused on double-negative tumors, which have been less well-studied. We found that many TFs previously linked to AR− mCRPC such as KLF5, MYCN, and FOXA2 were expressed at significantly higher levels in AR−/NE− tumors.
Figure 5.
Gene expression and DNA methylation analysis converges on KLF5 TF in AR−/NE− tumors. A, Heat map representing differentially expressed TFs in five subtypes of mCRPC. B, Heat map representing top 20 enriched TFs in hypomethylated regions of the five mCRPC subtypes. TFs are ranked by log (P value). The color intensity indicates the rank of the TFs from most enriched (dark red) to least enriched (white).
We next performed genome-wide differential methylation analysis comparing each subtype to AR+/NE− tumors, followed by motif enrichment analysis to identify TF binding sites that were preferentially exposed in that subtype. Hypomethylated regions in AR+/NE− tumors were enriched for motifs associated with Androgen Response elements, FOX family motifs, GRE motifs, and GRHL2 (Fig. 5B). This positive control result demonstrated differential methylation analysis could identify binding sites associated with driver TF and pioneer factors. Complementing these observations, hypermethylated regions in AR− tumors were enriched for ETS family motifs such as ETV2 and ERG, and androgen-associated motifs including HOXB13 and GRHL2 (Supplementary Fig. S7). Hypomethylated regions in AR−/NE+ tumors were significantly enriched for NE lineage-related TFs such as ASCL1 and NEUROD1 as well as TFs that promote epithelial-mesenchymal transition (EMT) including SNAIL1 and SLUG (Fig. 5B; refs. 63, 64).
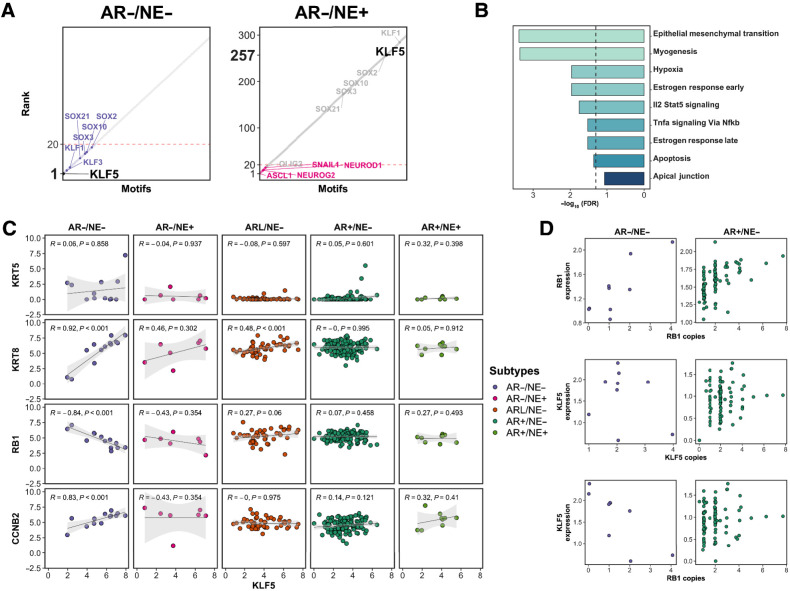
Focusing next on AR−/NE− tumors, we observed enrichment for motifs associated with SOX family and KLF motifs in the hypomethylated regions of this subtype (Fig. 5B). The KLF5 motif was the most highly enriched motif identified in AR−/NE− tumors, but it ranked 257th of 433 motifs in the AR−/NE+ subtype and was not enriched in AR+ subtypes (Fig. 6A; Supplementary Data 2). Among the KLF family genes with binding motifs enriched in AR−/NE− tumors (KLF5, KLF3, KLF1, KLF14, KLF6, KLF9), only KLF5 had significantly higher expression in AR−/NE− tumors (Fig. 5A; Supplementary Fig. S8). Genes harboring KLF5 binding sites that were hypomethylated in AR−/NE− tumors were enriched for roles in EMT, myogenesis, and estrogen response (Fig. 6B). This result was consistent with prior reports that KLF5 maintains epithelial cell identity in normal prostate and mammary tissues (65–67). To nominate subtype-specific associations between KLF5 and other genes linked to lineage phenotypes, we performed differential correlation analysis centered on KLF5. KLF5 expression was significantly correlated with luminal markers such as KRT18 in AR− and AR-low subtypes (Fig. 6C). KLF5 was not correlated with basal markers such as KRT5, which were expressed at low levels in all subtypes, though at significantly higher levels in AR−/NE− tumors than other subtypes (Fig. 6C). KLF5 expression levels were positively correlated with mitotic cyclin CCNB2 (Fig. 6C).
Figure 6.
Association between KLF5 TF enrichment and RB1 gene loss in AR−/NE− tumors. A, Rank order plots show the enrichment rank of KLF5 in AR−/NE− and AR−/NE+ subtypes on the left to right. Dashed red color indicates rank 20. B, Bar plots showing the gene set enrichment analyses for genes mapped to the KLF5 motif. Dashed line, FDR = 0.05. C, Scatter plots representing Spearman correlation between KLF5 gene expression and KRT5, KRT8, RB1, and CCNB2 genes. D, Scatter plots showing the relation between RB1 gene expression and RB1 copy numbers (top row), KLF5 gene expression and KLF5 copy numbers (middle row), and KLF5 gene expression and RB1 copy number (bottom row).
One of the strongest significant correlations we observed was an inverse correlation between expression of KLF5 and RB1 in AR−/NE− tumors (Figs. 6C and 4F). RB1 and KLF5 are located on chromosome 13 at 48.3 and 73 Mb, respectively. RB1 is frequently deleted in mCRPC (5), and expression levels of RB1 were correlated with RB1 copy number in both AR−/NE− and AR+/NE− tumors (Fig. 6D, top row). KLF5 was rarely deleted or amplified in AR−/NE− tumors, and there was no significant association between KLF5 expression and KLF5 copy number (Fig. 6D, middle row). KLF5 expression was, however, negatively correlated with RB1 copy levels only in AR−/NE− tumors (Fig. 6D, bottom row). The RB1 inverse correlation with KLF5 was the 22nd strongest correlation among all genes in the genome for AR−/NE− tumors. These observations were consistent with RB1 loss being linked to increased KLF5 activity in AR−/NE− tumors.
Discussion
Several studies have shown subtype heterogeneity among mCRPC tumors (5, 11, 13) and have identified that a subtype variously called small cell, neuroendocrine (7), t-SCNC (8), and aggressive variant (68) disease exists and has worse prognosis than prostate adenocarcinoma (8). This study characterized genomic and epigenomic drivers of mCRPC by integrating RNA-seq, deep WGS and WGBS, and clinical outcomes from 210 mCRPC tumors to assess subtypes defined by AR and NE status including adenocarcinoma (AR+/NE−), double-positive (AR+/NE+), low AR (ARL/NE−), neuroendocrine (AR−/NE+), and double-negative (AR−/NE−). We demonstrated that AR−/NE− tumors have the worst survival outcomes of these subtypes and harbor distinct genomic and epigenomic changes compared with the AR−/NE+ subtype, which may facilitate the identification of novel therapeutic targets in AR-independent tumors. We identified transcriptional subtypes that were consistent with five molecular subtypes reported by Labrecque and colleagues (13). These five subtypes were not observed in primary prostate tumors. This suggests mCRPC tumors evolve from the AR+/NE− phenotype concurrently with the development of castration-resistant disease in response to therapeutic pressure from androgen-targeting therapy (69). Our observation that patients with AR−/NE− tumors had the worst survival outcome among men with mCRPC who are actively being treated supports the expansion of the adenocarcinoma versus neuroendocrine dichotomy to include these five subtypes in genomic and clinical studies of mCRPC. The small number of tumors with the AR−/NE− phenotype in our cohort limited our statistical power to perform multivariate survival analysis.
AR−/NE− tumors were enriched for biallelic inactivation of PTEN and amplification of a DNA region that included CHD7, an ATP-dependent chromatin remodeling gene. Despite a low frequency of somatic changes in CHD7 among AR−/NE+ tumors, these tumors express the highest levels of CHD7. In normal tissues, CHD7 is abundantly expressed only in the cerebellum. CHD7 is essential for proper formation of the multipotent migratory neural crest (46), it plays an important role in promoting neural progenitor differentiation in ESCs, and it colocalizes with SOX2 (47–49). We identified two intragenic candidate enhancer regions of CHD7 (DMR2 and DMR3) that were hypomethylated in the AR−/NE+ subtype. Hypomethylation of DMR2 and DMR3 was significantly correlated with higher CHD7 expression, consistent with the profile of an enhancer. Published ChIP-seq experiments in neuroendocrine lineage tumors showed ASCL1 binds at DMR2 at the location of an ASCL1 binding motif. Analysis of chromatin interactions in models of prostate cancer using Chromatin Interaction Analysis with Paired-End Tag (ChIA-PET) techniques (70) would be informative to explore this relationship further; our observations predict influence of DMR2 and DMR3 would be conditional on whether the cells have a neuroendocrine phenotype. Ectopic overexpression of CHD7 in preclinical models of glioblastoma cell-line increases cell motility and invasiveness (71). Abundant prior evidence therefore links CHD7 to neural development, though to our knowledge this is the first study linking CHD7 to neuroendocrine mCRPC.
We nominated TFs specifically relevant to each subtype by unbiased genome-wide methylation analysis of TF binding motifs. This analysis underscored the profound differences in transcriptional control of AR−/NE− and AR−/NE+ tumor cells. ASCL1 binding motifs had the strongest enrichment in AR−/NE+ tumors. Together with our CHD7 analysis, this observation adds to emerging evidence that ASCL1 plays a key role driving lineage plasticity in this subtype (10). This analysis also showed KLF-family motifs were significantly enriched in the hypomethylated regions of AR−/NE− tumors. Among KLF family genes, KLF5 was most highly expressed in this subtype. A positive association has been reported between KLF5 gene expression and SPOP gene expression in an early-onset primary prostate tumors (72). It has been proposed that KLF5 plays contrasting roles in advanced prostate cancer depending on AR activity (65). In AR+ tumors, KLF5 interacts with AR and decreases AR expression. In the absence of AR, KLF5 has been reported to function as an oncogene that promotes cell migration and invasion (65). We observe highly divergent enrichment in our methylation analysis for KLF5 binding sites in AR−/NE− and AR−/NE+ subtypes. Notably, KLF5 was the most enriched motif in AR−/NE− tumors, while it ranked 257th in AR−/NE+ tumors. These observations, combined with elevated expression of KLF5 in AR−/NE− tumors, support our hypothesis that KLF5 drives AR−/NE− tumors. The link that we observed between elevated KLF5 expression and RB1 inactivation was striking, but further studies will be required to determine whether RB1 loss directly impacts KLF5 expression in AR− disease.
Supplementary Material
Clinical characteristics of samples in WCDT cohort
List of Differentially expressed genes in the subtypes of mCRPC in comparison with AR+/NE- tumors
Hierarchical clustering and principal component analyses (PCA) on the mCRPC tumors of WCDT cohort.
Molecular Charactersistcs of samples in WCDT cohort
List of enriched motifs identified in the hyper- and hypo-methylated regions of mCRPC subtypes
Hierarchical clustering analyses of benign, localized, and mCRPC tumors representing the expression of neuroendocrine marker genes (NE), androgen-related genes (AR), and genes related to squamous (SQUAM) differentiation
ETS family fusions detected in the WCDT samples
Volcano plots representing the differentially expressed (DE) genes in the mCRPC subtypes compared to AR+/NE- tumors
Gene Ontology (GO) with Biological processes (BP) on the genes uniquely up- or down-regulated within each subtype
Gene expression level of SOX2 among the tumor subtypes of WCDT samples
Gene expression level of CHD7 among the mCRPC tumors of Beltran et al. cohort
A heatmap representing the enrichment of transcription factors (TF) in hypermethylated regions of different subtypes of the WCDT samples
Gene expression level of Krüppel-like factor (KLF) family among the five mCRPC subtypes
Acknowledgments
The authors thank the patients who selflessly contributed samples to this study and without whom this research would not have been possible. This research was supported by a Stand Up To Cancer Prostate Cancer Foundation - Prostate Cancer Dream Team Translational Research grant (SU2C -AACR -DT0812). This research grant is made possible by the generous support of the Movember Foundation. Stand Up To Cancer is a division of the Entertainment Industry Foundation. This research grant was administered by the American Association for Cancer Research, the scientific partner of SU2C. R. Aggarwal, H. Li, and M. Sjöström were funded by Prostate Cancer Foundation (PCF) Young Investigator Awards. A. Lundberg was funded by a pilot grant from the UCSF Department of Urology. D.A. Quigley was funded by a Young Investigator and Challenge awards from the PCF and the UCSF Benioff Initiative for Prostate Cancer Research, and by the U.S. Department of Defense (W81XWH1910682). F.Y. Feng was funded by PCF Challenge Awards. Additional funding was provided by a UCSF Benioff Initiative for Prostate Cancer Research award. F.Y. Feng was supported by NIH/NCI 1R01CA230516–01, NIH/NCI 1R01CA227025, PCF 17CHAL06, and NIH P50CA186786.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Disclosures
R. Aggarwal reports personal fees from Dendreon, AAA/Novartis, Pfizer, Jubilant Therapeutics, Alessa, Exelixis, Bayer, Modra Therapeutics, Boxer Capital, Tersara, Deallus, EcoR1, Bioexcel, Targeted Oncology, Scitaris, OncLive; grants and personal fees from AstraZeneca, Janssen, Merck; and grants and personal fees from Amgen outside the submitted work. M. Sjöström reports grants from Prostate Cancer Foundation during the conduct of the study. J. Chou reports grants from Department of Defense Prostate Cancer Research Program, Prostate Cancer Foundation, and NCI; and personal fees from Exai Bio outside the submitted work. X. Zhu reports grants from Conquer Cancer Foundation and Prostate Cancer Foundation outside the submitted work. J.J. Alumkal reports other support from Astellas Pharma, Dendreon, Bristol Myers Squibb, Zenith Epigenetics, Beactica; and grants from Astellas Pharma/Pfizer outside the submitted work. T.M. Beer reports grants from Stand Up to Cancer/Prostate Cancer Foundation during the conduct of the study and other support from Exact Sciences outside the submitted work. K.N. Chi reports grants from Stand Up to Cancer during the conduct of the study; grants and personal fees from AstraZeneca, Bayer, Merck, Novartis, Pfizer, Point Biopharma, Roche; personal fees from Astellas; and grants and personal fees from Janssen outside the submitted work. C.P. Evans reports grants and personal fees from Astellas, Janssen, Janssen, and Anchiano Therapeutics outside the submitted work. M.B. Rettig reports grants and nonfinancial support from Novartis, Progenics; nonfinancial support from Merck; personal fees from Janssen, INmune Bio, Bayer, Myovant, Ambrx, Amgen, and Roivant-Oncopeia outside the submitted work; in addition, M.B. Rettig has a patent for Inhibtors of the N-terminus of the Androgen Receptor issued. O.N. Witte reports grants from SU2C PCF West Coast Dream Team during the conduct of the study and other support from various companies outside the submitted work. A.W. Wyatt reports personal fees from AstraZeneca, Merck, Janssen, Bayer, Pfizer; grants from ESSA Pharma; and personal fees from EMD Serono outside the submitted work. F.Y. Feng reports other support from Artera, ClearNote, SerImmune; personal fees from BMS, Janssen, Roivant, Myovant, Astellas, Sanofi, BlueEarth Diagnostics, and Tempus outside the submitted work. No disclosures were reported by the other authors.
Authors' Contributions
A. Lundberg: Conceptualization, resources, data curation, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. M. Zhang: Conceptualization, data curation, writing–original draft, writing–review and editing. R. Aggarwal: Resources, data curation, writing–review and editing. H. Li: Resources, validation, writing–review and editing. L. Zhang: Data curation, writing–review and editing. A. Foye: Resources, methodology, writing–review and editing. M. Sjöström: Conceptualization, data curation, methodology, writing–review and editing. J. Chou: Resources, methodology, writing–review and editing. K. Chang: Validation, methodology, writing–review and editing. T. Moreno-Rodriguez: Data curation, writing–review and editing. R. Shrestha: Data curation, writing–review and editing. A. Baskin: Writing–review and editing. X. Zhu: Conceptualization, writing–review and editing. A.S. Weinstein: Writing–review and editing. N. Younger: Writing–review and editing. J.J. Alumkal: Resources, writing–review and editing. T.M. Beer: Resources, writing–review and editing. K.N. Chi: Resources, writing–review and editing. C.P. Evans: Resources, writing–review and editing. M. Gleave: Resources, writing–review and editing. P.N. Lara: Resources, writing–review and editing. R.E. Reiter: Resources, writing–review and editing. M.B. Rettig: Resources, writing–review and editing. O.N. Witte: Resources, writing–review and editing. A.W. Wyatt: Resources, methodology, writing–review and editing. F.Y. Feng: Conceptualization, resources, data curation, supervision, funding acquisition, investigation, methodology, project administration, writing–review and editing. E.J. Small: Conceptualization, resources, investigation, methodology, project administration, writing–review and editing. D.A. Quigley: Conceptualization, resources, supervision, funding acquisition, investigation, project administration, writing–review and editing.
References
- 1. Cancer facts & figures 2022 [Internet]. American Cancer Society, Atlanta, Ga; 2022. Available from:https://www.cancer.org/cancer/prostate-cancer/detection-diagnosis-staging/survival-rates.html. [Google Scholar]
- 2. Watson PA, Arora VK, Sawyers CL. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat Rev Cancer 2015;15:701–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Taplin M-E, Bubley GJ, Shuster TD, Frantz ME, Spooner AE, Ogata GK, et al. Mutation of the androgen-receptor gene in metastatic androgen-independent prostate cancer. N Engl J Med 1995;332:1393–8. [DOI] [PubMed] [Google Scholar]
- 4. Robinson D, Van Allen EM, Wu Y-M, Schultz N, Lonigro RJ, Mosquera J-M, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Quigley DA, Dang HX, Zhao SG, Lloyd P, Aggarwal R, Alumkal JJ, et al. Genomic hallmarks and structural variation in metastatic prostate cancer. Cell 2018;174:758–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Herberts C, Annala M, Sipola J, Ng SWS, Chen XE, Nurminen A, et al. Deep whole-genome ctDNA chronology of treatment-resistant prostate cancer. Nature 2022;608:199–208. [DOI] [PubMed] [Google Scholar]
- 7. Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J, et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med 2016;22:298–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Aggarwal R, Huang J, Alumkal JJ, Zhang L, Feng FY, Thomas GV, et al. Clinical and genomic characterization of treatment-emergent small-cell neuroendocrine prostate cancer: a multi-institutional prospective study. J Clin Oncol 2018;36:2492–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Aggarwal RR, Quigley DA, Huang J, Zhang L, Beer TM, Rettig MB, et al. Whole-genome and transcriptional analysis of treatment-emergent small-cell neuroendocrine prostate cancer demonstrates intraclass heterogeneity. Mol Cancer Res 2019;17:1235–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nouruzi S, Ganguli D, Tabrizian N, Kobelev M, Sivak O, Namekawa T, et al. ASCL1 activates neuronal stem cell–like lineage programming through remodeling of the chromatin landscape in prostate cancer. Nat Commun 2022;13:2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhao SG, Chen WS, Li H, Foye A, Zhang M, Sjöström M, et al. DNA methylation landscapes in advanced prostate cancer. Nat Genet 2020;52:778–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sjöström M, Zhao SG, Levy S, Zhang M, Ning Y, Shrestha R, et al. The 5-Hydroxymethylcytosine landscape of prostate cancer. Cancer Res 2022;82:3888–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Labrecque MP, Coleman IM, Brown LG, True LD, Kollath L, Lakely B, et al. Molecular profiling stratifies diverse phenotypes of treatment-refractory metastatic castration-resistant prostate cancer. J Clin Invest 2019;129:4492–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Priestley P, Baber J, Lolkema MP, Steeghs N, de Bruijn E, Shale C, et al. Pan-cancer whole-genome analyses of metastatic solid tumors. Nature 2019;575:210–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 2013;31:213–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Callari M, Sammut S-J, De Mattos-Arruda L, Bruna A, Rueda OM, Chin S-F, et al. Intersect-then-combine approach: improving the performance of somatic variant calling in whole exome sequencing data using multiple aligners and callers. Genome Med 2017;9:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cameron DL, Baber J, Shale C, Papenfuss AT, Valle-Inclan JE, Besselink N, et al. GRIDSS, PURPLE, LINX: unscrambling the tumor genome via integrated analysis of structural variation and copy number [Internet]. Bioinformatics; 2019. Available from:http://biorxiv.org/lookup/doi/10.1101/781013.
- 20. Wu H, Xu T, Feng H, Chen L, Li B, Yao B, et al. Detection of differentially methylated regions from whole-genome bisulfite sequencing data without replicates. Nucleic Acids Res 2015;43:e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Castro-Mondragon JA, Riudavets-Puig R, Rauluseviciute I, Lemma RB, Turchi L, Blanc-Mathieu R, et al. JASPAR 2022: the 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res 2022;50:D165–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics 2011;27:1017–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Amemiya HM, Kundaje A, Boyle AP. The ENCODE blacklist: identification of problematic regions of the genome. Sci Rep. 2019;9:9354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 2010;38:576–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf 2013;14:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. The Innovation 2021;2:100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 2016;44:W90–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fraser M, Sabelnykova VY, Yamaguchi TN, Heisler LE, Livingstone J, Huang V, et al. Genomic hallmarks of localized, non-indolent prostate cancer. Nature 2017;541:359–64. [DOI] [PubMed] [Google Scholar]
- 29. Pinskaya M, Saci Z, Gallopin M, Gabriel M, Nguyen HT, Firlej V, et al. Reference-free transcriptome exploration reveals novel RNAs for prostate cancer diagnosis. Life Sci Alliance 2019;2:e201900449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Meuleman W, Muratov A, Rynes E, Halow J, Lee K, Bates D, et al. Index and biological spectrum of human DNase I hypersensitive sites. Nature 2020;584:244–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stelloo S, Nevedomskaya E, Kim Y, Schuurman K, Valle-Encinas E, Lobo J, et al. Integrative epigenetic taxonomy of primary prostate cancer. Nat Commun 2018;9:4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Taavitsainen S, Engedal N, Cao S, Handle F, Erickson A, Prekovic S, et al. Single-cell ATAC and RNA sequencing reveal preexisting and persistent cells associated with prostate cancer relapse. Nat Commun 2021;12:5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Su W, Han HH, Wang Y, Zhang B, Zhou B, Cheng Y, et al. The polycomb repressor complex 1 drives double-negative prostate cancer metastasis by coordinating stemness and immune suppression. Cancer Cell 2019;36:139–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bluemn EG, Coleman IM, Lucas JM, Coleman RT, Hernandez-Lopez S, Tharakan R, et al. Androgen receptor pathway-independent prostate cancer is sustained through FGF signaling. Cancer Cell 2017;32:474–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Aparicio A, Logothetis CJ, Maity SN. Understanding the lethal variant of prostate cancer: power of examining extremes. Cancer Discov 2011;1:466–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. PCF/SU2C International Prostate Cancer Dream Team, Armenia J, Wankowicz SAM, Liu D, Gao J, Kundra R, et al. The long tail of oncogenic drivers in prostate cancer. Nat Genet 2018;50:645–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ku SY, Rosario S, Wang Y, Mu P, Seshadri M, Goodrich ZW, et al. Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science 2017;355:78–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Carver BS, Chapinski C, Wongvipat J, Hieronymus H, Chen Y, Chandarlapaty S, et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell 2011;19:575–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chen Y, Chi P, Rockowitz S, Iaquinta PJ, Shamu T, Shukla S, et al. ETS factors reprogram the androgen receptor cistrome and prime prostate tumorigenesis in response to PTEN loss. Nat Med 2013;19:1023–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ferraldeschi R, Nava Rodrigues D, Riisnaes R, Miranda S, Figueiredo I, Rescigno P, et al. PTEN protein loss and clinical outcome from castration-resistant prostate cancer treated with abiraterone acetate. Eur Urol 2015;67:795–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Castro E, Goh C, Olmos D, Saunders E, Leongamornlert D, Tymrakiewicz M, et al. Germline BRCA mutations are associated with higher risk of nodal involvement, distant metastasis, and poor survival outcomes in prostate cancer. J Clin Oncol 2013;31:1748–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kaufman B, Shapira-Frommer R, Schmutzler RK, Audeh MW, Friedlander M, Balmaña J, et al. Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J Clin Oncol 2015;33:244–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mateo J, Carreira S, Sandhu S, Miranda S, Mossop H, Perez-Lopez R, et al. DNA-repair defects and olaparib in metastatic prostate cancer. N Engl J Med 2015;373:1697–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Qiu X, Boufaied N, Hallal T, Feit A, de Polo A, Luoma AM, et al. MYC drives aggressive prostate cancer by disrupting transcriptional pause release at androgen receptor targets. Nat Commun 2022;13:2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Barfeld SJ, Urbanucci A, Itkonen HM, Fazli L, Hicks JL, Thiede B, et al. c-Myc antagonises the transcriptional activity of the androgen receptor in prostate cancer affecting key gene networks. EBioMedicine 2017;18:83–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bajpai R, Chen DA, Rada-Iglesias A, Zhang J, Xiong Y, Helms J, et al. CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature 2010;463:958–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schnetz MP, Handoko L, Akhtar-Zaidi B, Bartels CF, Pereira CF, Fisher AG, et al. CHD7 targets active gene enhancer elements to modulate ES cell-specific gene expression. van Heyningen V, editor. PLoS Genet 2010;6:e1001023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Engelen E, Akinci U, Bryne JC, Hou J, Gontan C, Moen M, et al. Sox2 cooperates with Chd7 to regulate genes that are mutated in human syndromes. Nat Genet 2011;43:607–11. [DOI] [PubMed] [Google Scholar]
- 49. Yao H, Hannum DF, Zhai Y, Hill SF, ’Oliveira ARD, Lou W, et al. CHD7 promotes neural progenitor differentiation in embryonic stem cells via altered chromatin accessibility and nascent gene expression. Sci Rep 2020;10:17445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mu P, Zhang Z, Benelli M, Karthaus WR, Hoover E, Chen C-C, et al. SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer. Science 2017;355:84–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Grimm D, Bauer J, Wise P, Krüger M, Simonsen U, Wehland M, et al. The role of SOX family members in solid tumors and metastasis. Semin Cancer Biol 2020;67:122–53. [DOI] [PubMed] [Google Scholar]
- 52. Zhong W, Qin G, Dai Q, Han Z, Chen S, Ling X, et al. SOXs in human prostate cancer: implication as progression and prognosis factors. BMC Cancer 2012;12:248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Suelves M, Carrió E, Núñez-Álvarez Y, Peinado MA. DNA methylation dynamics in cellular commitment and differentiation. Brief Funct Genomics 2016;15:443–53. [DOI] [PubMed] [Google Scholar]
- 54. Hashimshony T, Zhang J, Keshet I, Bustin M, Cedar H. The role of DNA methylation in setting up chromatin structure during development. Nat Genet 2003;34:187–92. [DOI] [PubMed] [Google Scholar]
- 55. Shlyueva D, Stampfel G, Stark A. Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet 2014;15:272–86. [DOI] [PubMed] [Google Scholar]
- 56. Aran D, Sabato S, Hellman A. DNA methylation of distal regulatory sites characterizes dysregulation of cancer genes. Genome Biol 2013;14:R21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Becker PB, Ruppert S, Schütz G. Genomic footprinting reveals cell type-specific DNA binding of ubiquitous factors. Cell 1987;51:435–43. [DOI] [PubMed] [Google Scholar]
- 58. Weih F, Nitsch D, Reik A, Schütz G, Becker PB. Analysis of CpG methylation and genomic footprinting at the tyrosine aminotransferase gene: DNA methylation alone is not sufficient to prevent protein binding in vivo. EMBO J 1991;10:2559–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Maurano MT, Wang H, John S, Shafer A, Canfield T, Lee K, et al. Role of DNA methylation in modulating transcription factor occupancy. Cell Rep 2015;12:1184–95. [DOI] [PubMed] [Google Scholar]
- 60. Lennon MJ, Jones SP, Lovelace MD, Guillemin GJ, Brew BJ. Bcl11b–A critical neurodevelopmental transcription factor–Roles in health and disease. Front Cell Neurosci 2017;11:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tsigelny IF, Kouznetsova VL, Lian N, Kesari S. Molecular mechanisms of OLIG2 transcription factor in brain cancer. Oncotarget 2016;7:53074–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lambert SA, Jolma A, Campitelli LF, Das PK, Yin Y, Albu M, et al. The human transcription factors. Cell 2018;172:650–65. [DOI] [PubMed] [Google Scholar]
- 63. Yu Q, Zhang K, Wang X, Liu X, Zhang Z. Expression of transcription factors snail, slug, and twist in human bladder carcinoma. J Exp Clin Cancer Res 2010;29:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wang J, He C, Gao P, Wang S, Lv R, Zhou H, et al. HNF1B-mediated repression of SLUG is suppressed by EZH2 in aggressive prostate cancer. Oncogene 2020;39:1335–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Che M, Chaturvedi A, Munro SA, Pitzen SP, Ling A, Zhang W, et al. Opposing transcriptional programs of KLF5 and AR emerge during therapy for advanced prostate cancer. Nat Commun 2021;12:6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Liu R, Shi P, Zhou Z, Zhang H, Li W, Zhang H, et al. Krüpple-like factor 5 is essential for mammary gland development and tumorigenesis. J Pathol 2018;246:497–507. [DOI] [PubMed] [Google Scholar]
- 67. Zhang B, Li Y, Wu Q, Xie L, Barwick B, Fu C, et al. Acetylation of KLF5 maintains EMT and tumorigenicity to cause chemoresistant bone metastasis in prostate cancer. Nat Commun 2021;12:1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Aparicio AM, Shen L, Tapia ELN, Lu J-F, Chen H-C, Zhang J, et al. Combined tumor suppressor defects characterize clinically defined aggressive variant prostate cancers. Clin Cancer Res 2016;22:1520–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Westbrook TC, Guan X, Rodansky E, Flores D, Liu CJ, Udager AM, et al. Transcriptional profiling of matched patient biopsies clarifies molecular determinants of enzalutamide-induced lineage plasticity. Nat Commun 2022;13:5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ramanand SG, Chen Y, Yuan J, Daescu K, Lambros MBK, Houlahan KE, et al. The landscape of RNA polymerase II–associated chromatin interactions in prostate cancer. J Clin Invest 2020;130:3987–4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Machado RAC, Schneider H, DeOcesano-Pereira C, Lichtenstein F, Andrade F, Fujita A, et al. CHD7 promotes glioblastoma cell motility and invasiveness through transcriptional modulation of an invasion signature. Sci Rep 2019;9:3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Gerhauser C, Favero F, Risch T, Simon R, Feuerbach L, Assenov Y, et al. Molecular evolution of early-onset prostate cancer identifies molecular risk markers and clinical trajectories. Cancer Cell 2018;34:996–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clinical characteristics of samples in WCDT cohort
List of Differentially expressed genes in the subtypes of mCRPC in comparison with AR+/NE- tumors
Hierarchical clustering and principal component analyses (PCA) on the mCRPC tumors of WCDT cohort.
Molecular Charactersistcs of samples in WCDT cohort
List of enriched motifs identified in the hyper- and hypo-methylated regions of mCRPC subtypes
Hierarchical clustering analyses of benign, localized, and mCRPC tumors representing the expression of neuroendocrine marker genes (NE), androgen-related genes (AR), and genes related to squamous (SQUAM) differentiation
ETS family fusions detected in the WCDT samples
Volcano plots representing the differentially expressed (DE) genes in the mCRPC subtypes compared to AR+/NE- tumors
Gene Ontology (GO) with Biological processes (BP) on the genes uniquely up- or down-regulated within each subtype
Gene expression level of SOX2 among the tumor subtypes of WCDT samples
Gene expression level of CHD7 among the mCRPC tumors of Beltran et al. cohort
A heatmap representing the enrichment of transcription factors (TF) in hypermethylated regions of different subtypes of the WCDT samples
Gene expression level of Krüppel-like factor (KLF) family among the five mCRPC subtypes
Data Availability Statement
RNA-seq FASTQ files of 148 localized samples from the CPC-GENE cohort (28) were obtained from the European Genome-Phenome Archive (EGA) under accession number EGAS00001000900 and the FASTQ files of eight benign samples from the PAIR cohort (29) were retrieved from Gene Expression Omnibus (GEO) database under accession number GSE115414. The files were aligned with STAR, and the gene level quantification was performed using gene models in GENECODE version 28. The expression value of each gene was converted to TPM. The chromatin immunoprecipitation sequencing (ChIP-seq) data of DNase I hypersensitive sites (DHS; ref. 30) was obtained from the ENCODE project under accession number ENCSR857UZV. The H3K27ac ChIP-seq data of primary prostate tumors (31) was obtained from GEO, under accession number GSE120738. WGBS and WGS from 100 samples of mCRPC tumors from the West Coast Prostate Cancer Dream Team (WCDT) cohort are available on dbGaP with study number phs001648 (11) and an additional 28 samples are available on EGA with study number EGAS00001006649. RNA-seq data from 210 samples of mCRPC tumors from the WCDT cohort are available on EGA with study numbers EGAD00001008991, EGAD00001008487, and EGAD00001009065 (Supplementary Table S2). All other raw data are available upon request from the corresponding author.