Abstract
PARP inhibitors (PARPi) are a kind of cancer therapy that targets poly (ADP-ribose) polymerase. PARPi is the first clinically approved drug to exert synthetic lethality by obstructing the DNA single-strand break repair process. Despite the significant therapeutic effect in patients with homologous recombination (HR) repair deficiency, innate and acquired resistance to PARPi is a main challenge in the clinic. In this review, we mainly discussed the underlying mechanisms of PARPi resistance and summarized the promising solutions to overcome PARPi resistance, aiming at extending PARPi application and improving patient outcomes.
Keywords: Drug resistance, Homologous recombination, PARP, PARP inhibitor, Poly (ADP-ribose) polymerase
Introduction
Poly (ADP-ribose) polymerase (PARP) is a multifunctional post-translational modification enzyme, which participates in various biological processes.1 Among all the 17 members of the poly (ADP-ribose) polymerases (PARPs) family, PARP1 is a particularly important protein for DNA damage repair and genome stability, which catalyzes the covalent attachment of PAR polymers on itself and other specific proteins, including histones, DNA repair proteins, and chromatin modulators using NAD+ as a donor of ADP-ribose units.2, 3, 4 PARP1 contains several domains: N-terminal zinc finger motif, BRCT domain, WGR domain, and C-terminal catalytic (CAT) domain.5 PARP1 plays an essential role in repairing single-strand breaks (SSBs).6 Since SSBs are an intermediate of base-excision repair (BER),7 PARP1 is also required for BER.8 Once DNA damage occurs, PARP1 is activated and initiates the auto-PARylation, which further activates PARP1 and enables the PARylation of histones and other DNA damage-associated proteins. Eventually, this auto- and hetero-modification recruit the downstream DNA repair proteins, such as XRCC1, to the DNA damage site, promoting the effective repair of DNA.9,10
PARP1 is also cleaved between Asp124 and Gly215 by caspases, core members of apoptosis,11 resulting in two specific segments: the catalytic domain (89 kD) of the carboxyl-terminal fragment and DNA-binding domain (24 kD) of the amino-terminal segment.11 In addition to the role of PARP1 in DNA damage repair and apoptosis, PARP1 also regulates the activity of RNA polymerase II by inhibiting the negative elongation factor (NELF) and modulating the gene transcription process.12
The homologous recombination (HR) repair pathway is a major pathway to repair the DNA double-strand breaks (DSBs), and HR deficiency leads to genomic instability and tumorigenesis.13 PARP inhibitor (PARPi), which inhibits DNA SSBs repair, exerts a synthetic lethality with tumor-specific HR deficiency and presents the anti-tumor effect. Emerging evidence supports the anti-tumor role of PARPi in both inherited cancer with BRCA mutation, and in sporadic cancer harboring HR repair deficiency.14 Synergistic effects were observed in the combination treatment with PARPi and platinum-based chemotherapy. Platinum can covalently crosslink DNA to cause DSBs, while PARPi can inhibit DNA single-strand repair.15 Therefore, patients with platinum-sensitive tumors are more likely to benefit from the therapy of PARPi. Even in patients with BRCA mutations, the efficacy of PARPi is still related to the response to platinum.16 Although PARPi is a promising treatment, the majority of patients will relapse due to acquired resistance. PARPi resistance is becoming a challenge, threatening the efficacy of PARPi. There is a critical need to identify the molecular mechanisms of resistance to PARPi and explore strategies to overcome the resistance.
The mechanism of action of PARPi
Since 2014, when olaparib (AZD2281) was granted accelerated approval by the United States Food and Drug Administration (FDA) as monotherapy in inherited BRCA-mutated ovarian cancer,17,18 PARP inhibitors, including olaparib, rucaparib, niraparib, and talazoparib, have received approvals by FDA to be used in various cancers based on their revolutionary results in clinical trials (Fig. 1). PARP1, the most extensively studied PARP member, will be used as an example to illustrate the mechanisms of PARPi.
Figure 1.
Hallmarks in the field of PARP inhibitors.
PARP1 catalytic inhibition
PARP1 is recruited to the site of SSBs and its C-terminal catalytic (CAT) domain can be rapidly activated to hydrolyze NAD+, leading to the PARylation of several proteins as well as itself,19,20 which initiates DNA repair mechanisms (Fig. 2). This binding of NAD+ to PARP1 recruits DNA repair proteins to the site of DNA damage.21 PARP inhibitor is a nicotinamide analogue bearing the nicotinamide moiety that could compete with NAD+ to bind PARP1.22 PARPi kills cancer cells by blocking the synthesis of PAR chains and interfering with the repair of SSBs. PARPi does not affect the condensation level of undamaged DNA but blocks the reversal of condensation of damaged DNA in the presence of NAD+.23
Figure 2.
The mechanism of action of PARPi. PARPs are recruited to the sites of DNA single-strand breaks (SSBs) and rapidly activated, which initiate and maintain DNA damage repair. PARPi contains a nicotinamide moiety that binds PARPs, which blocks the synthesis of PAR chains and interferes with the repair of SSBs. PARP1 autoPARylation leads to its release from the site of DNA damage. After the competitive binding of PARPi, PARPs are allosteric and enhance the binding strength to damaged DNA. Trapped PARPs lead to the accumulation of unrepaired SSBs, which impair the proper progression of replication forks and ultimately result in the formation of DSBs that need homologous recombination (HR) repair. If the HR repair is deficient, the DNA breaks might not be repaired and ultimately lead to cell death.
PARP1 trapping
PARPi also works through PARP1 trapping. In general, PARP1 autoPARylation induces itself to release from the DNA damage site.24 After the competitive binding of PARPi, PARP1 is allosteric, which enhances PARP1 binding to damaged DNA.25 The trapped PARP1 on SSBs encounters replication forks and then leads to DSBs that need to be repaired by HR.26 If PARPi is used on HR-deficient cells, the DNA breaks could not be repaired, which may impede replication forks and ultimately lead to cell death.27 Thus, trapped PARP1-DNA complexes are more cytotoxic than unrepaired SSBs caused by PARP1 inactivation. After being captured, PARP1 is SUMOylated by PIAS4 (protein inhibitor of activated STAT 4) and subsequently ubiquitylated by the Sumo-targeted E3 Ubiquitin ligase RNF4, promoting recruitment of P97 and removal of trapped PARP1 from chromatin. P97 complex inhibitors prolong PARP1 trapping and enhance PARPi-related cytotoxicity in HR-deficient tumor cells.28 The elevated repairability of PARP-DNA complexes is linked to acquired drug resistance.
Replication gap
The replication gap is another key determinant of the mortality of PARPi in BRCA defective cells. BRCA1-or FANCJ-deficient cells exhibit common repair deficiencies and distinct PARPi responses, implying that the replication gap might be a key differentiating factor. The sensitivity to PARPi aligns with the extent of replication gap formation, and replication gaps in BRCA1-deficient cells are caused by Okazaki fragment processing (OFP) defects. Targeting gaps can resensitize and augment PARPi synthetic lethality.29 Spartan (SPRTN), a metalloprotease involved in DNA replication, is associated with the response to various PARP inhibitors. SPRTN-deficient cells were sensitive to olaparib and talazoparib since SPRTN-deficient cells showed delayed clearance of trapped PARP1 and replication fork delay when treated with talazoparib or olaparib.30 However, SPRTN-deficient cells were not sensitive to PARP trapper veliparib.
Current status of PARPi drugs
Olaparib is the first PARPi validated in clinical trials, which demonstrated its efficacy and safety in several malignancies. Thus far, olaparib, niraparib, talazoparib, and rucaparib have been approved for selected patients with breast, ovarian, or pancreatic cancer.31,32 The timeline for the development of PARPi is shown in Figure 1, and the clinical progress of several major PARP inhibitors is summarized in Table 1.
Table 1.
Clinical progress of several major PARP inhibitors.
| Drug | Types of cancer | Approval (FDA/EMA/NMPA) | Ongoing clinical trials |
|||
|---|---|---|---|---|---|---|
| Clinical trials | Phase | Therapy | Inclusion criteria | |||
| Olaparib | Ovarian cancer |
|
NCT03737643 | 3 | Durvalumab + bevacizumab + olaparib after durvalumab + chemotherapy + bevacizumab | Newly diagnosed advanced ovarian cancer |
| Breast cancer |
|
NCT03150576 | 3 | Olaparib + platinum | TNBC in the neoadjuvant setting | |
| Pancreatic cancer | Metastatic gBRCA-mut pancreatic adenocarcinoma | |||||
| Prostate cancer | Metastatic castration-resistant prostate cancer with HRR gene-mutated. | NCT03732820 | 3 | Olaparib + abiraterone + prednisone | mCRPC | |
| Rucaparib | Ovarian cancer | Metastatic ovarian cancer with gBRCA mutations. | NCT03522246 | 3 | Rucaparib + nivolumab | Metastatic ovarian cancer sensitive to platinum |
| NCT04227522 | 3 | Maintenance therapy | Metastatic ovarian cancer | |||
| NCT02855944 | 3 | Rucaparib | Metastatic ovarian cancer | |||
| Prostate cancer | Metastatic castration-resistant prostate cancer with BRCA-mut | NCT02975934 | 3 | Rucaparib | Metastatic castration-resistant prostate cancer with HRD | |
| NCT04455750 | 3 | Rucaparib + enzalutamide | Metastatic testosterone-deprivation-resistant prostate cancer | |||
| Niraparib | Ovarian cancer |
|
NCT05460000 | 3 | Niraparib maintenance after chemotherapy | Advanced ovarian cancer with HRD |
| NCT03705156 | 3 | Niraparib maintenance after chemotherapy | Advanced platinum-sensitive ovarian cancer with HRD | |||
| NCT03598270 | 3 | Niraparib maintenance with/without atezolizumab after chemotherapy with/without atezolizumab | Advanced ovarian cancer | |||
| NCT05009082 | 3 | Niraparib with/without bevacizumab | Advanced ovarian cancer | |||
| Breast cancer | NCT04915755 | 3 | Niraparib | Early BRCA-mut HER2 negative breast cancer with and TNBC with detected ctDNA | ||
| Prostate cancer | NCT04497844 | 3 | Niraparib + abiraterone acetate + prednisone | Metastatic castration-sensitive prostate cancer with HRR gene-mutated | ||
| NCT03748641 | 3 | Niraparib + abiraterone acetate + prednisone | Metastatic prostate cancer | |||
| Non-small cell lung cancer | NCT04475939 | 3 | Niraparib + pembrolizumab as maintenance therapy | Stage IIIB/IIIC or IV non-small cell lung cancer sensitive to platinum and pembrolizumab | ||
| Talazoparib | Breast cancer | Advanced gBRCA mutated HER2-negative breast cancer | ||||
| Ovarian cancer | NCT03642132 | 3 | Avelumab + talazoparib as maintenance therapy | Newly diagnosed locally advanced or metastatic ovarian cancer | ||
| Prostate cancer | NCT04821622 | 3 | Talazoparib + enzalutamide | DDR-deficient mCSPC | ||
| NCT03395197 | 3 | Talazoparib + enzalutamide | Metastatic castration-resistant prostate cancer | |||
| Fuzuloparib (NMPA) | Ovarian cancer | advanced BRCA-mut platinum-sensitive ovarian cancer | ||||
| Veliparib | Breast cancer | NCT02163694 (has results) | 3 | Carboplatin + paclitaxel + veliparib | HER2-negative BRCA-associated breast cancer | |
| Ovarian cancer | NCT02470585 | 3 | Veliparib + carboplatin + paclitaxel | Advanced ovarian cancer | ||
| IMP4927 | Ovarian cancer | NCT04169997 | 3 | IMP4297 | Advanced ovarian cancer | |
| Pamiparib | Ovarian cancer | NCT03519230 | 3 | Pamiparib | Platinum-sensitive recurrent ovarian cancer | |
| AZD5305 | Prostate Cancer | NCT05367440 | I/IIa | AZD5305 + new hormonal agents | Metastatic prostate cancer | |
| solid tumors | NCT04644068 | I/IIa | AZD5305 with/without anti-cancer agents | Advanced solid malignancies | ||
Note: A platinum-sensitive tumor is defined as a tumor that does not recur within 6 months or more following platinum-based therapy. BRCA, breast cancer gene; DDR, DNA damage response; EMA, European Medicines Agency; FDA, Food and Drug Administration; NMPA, National Medical Products Administration; gBRCA, germline BRCA; HER2, human epidermal growth factor receptor 2; HRD, homologous recombination deficiency; HRR, homologous recombination repair; mCSPC, metastatic castration-resistant prostate cancer.
Olaparib
In 2009, the first clinical trial of the PARPi demonstrated a synthetic lethal for olaparib in breast cancer with BRCA1/BRCA2 deficiency.33 As more clinical trials have demonstrated its efficacy and safety, olaparib has been approved for pancreatic cancer, breast, and ovarian with BRCA1/2 mutations. A recent 5-year follow-up data from a randomized, double-blinded, phase III trial (SOLO1/GOG 3004) showed that the benefits of 2 years of olaparib maintenance continued until the end of treatment with newly diagnosed advanced ovarian cancer with BRCA mutations. Median progression-free survival (PFS) is prolonged to 4.5 years.34 SOLO2 trial also demonstrated that olaparib significantly improved overall survival (OS) and PFS as maintenance therapy in platinum-sensitive recurrent ovarian cancer (PROC) patients with BRCA mutations. However, post-hoc analyses suggested that patients who progressed after olaparib had less benefit from subsequent platinum-based chemotherapy than those who did not receive PARPi. Time to second progression was significantly longer in the placebo group than that in the olaparib group (12.1 months vs. 6.9 months).35 In clinical trials of metastatic castration-resistant prostate cancer (mCRPC), they revealed the synergy effect of olaparib combined with cabazitaxel in mCRPC patients who relapsed after docetaxel and androgen receptor axis targeted therapy.36 Furthermore, olaparib showed a longer PFS and a higher response rate.36,37 The exploratory analysis of the TOPARP-B trial showed that prostate cancer patients with homozygous loss of BRCA2, PALB2, and ATM derived more benefit from the addition of olaparib.38 For the mCRPC patients with DNA-repair defects who failed prior hormonal therapy, olaparib monotherapy also showed higher response rates and longer PFS compared to enzalutamide or abiraterone.39,40 A recent study suggests that olaparib maintenance therapy could bring survival benefits to metastatic pancreatic cancer patients with germline BRCA1/2 (gBRCA1/2) mutations.41
Rucaparib
Rucaparib maintenance is a safe and effective therapy for platinum-sensitive advanced pancreatic cancer with BRCA1/2 or PALB2 pathogenic variant.42 ARIEL2, a single-arm phase II trial, revealed the efficacy and safety of rucaparib in relapsed, high-grade ovarian cancer.43 It also proved that RAD51C and RAD51D mutations, as well as hypermethylation of BRCA1 promotors, were associated with better responses to rucaparib. The loss of BRCA1 methylation may be the main mechanism of cross-resistance between platinum and PARPi.43
Niraparib
De novo advanced ovarian cancer patients who received niraparib had significantly longer PFS regardless of the presence of HR defects.44 Niraparib is a useful option for maintenance therapy for relapse ovarian cancer sensitive to platinum.45 Additionally, niraparib is shown to significantly prolong PFS with manageable toxicity for patients with platinum-sensitive, extensive-stage small cell lung cancer (ES-SCLC) when used for maintenance therapy.46
Talazoparib
Talazoparib got approval for the treatment of HER2-negative metastatic breast cancer with gBRCA1/2 mutations due to its efficacy and safety.47 Talazoparib showed durable anti-tumor activity in patients with heavily pretreated advanced anti-castration prostate cancer altered by the DNA damage response (DDR)-HR genes.48
Fluzoparib
In a single-arm, phase II study, fluzoparib showed a good anti-tumor activity with an acceptable safety profile for patients with platinum-sensitive recurrent ovarian cancer with gBRCA1/2 mutations.49 A phase III FZOCUS-2 trial demonstrated a significant improvement in PFS with the utility of fluzoparib for maintenance therapy in platinum-sensitive, relapsed ovarian cancer patients, regardless of BRCA1/2 status.50
AZD5305
Studies have shown that the main synthetic lethal effect together with BRCA mutation is PARP1, suggesting that the inhibition effect on PARP2 may not be necessary to play a therapeutic role.51,52 Some PARP inhibitors failed in combination with other chemotherapies mainly owing to overlapping hematotoxicity, which is caused by its effects on PARP2 and other PARPs. Thus, finding a highly selective PARP1 inhibitor with reduced toxicity caused by cross-inhibition of PARP2 has become one of the important research directions for scientists. In 2015, Nerviano Medical Sciences first reported an effective, oral, and highly selective PARP1 inhibitor, NMS-P118, with good ADME (absorption, distribution, metabolism, and excretion).53 The pace of research has never stopped. AZD5305, a new generation of PARP1 specific inhibitor, is also a PARP1-DNA trapper, which showed good efficacy in vivo using the BRCA mutant HBC-17 patient-derived tumor xenograft (PDX) model.54 Giuditta et al showed that AZD5305 was 500-fold more selective for PARP1 over PARP2. Surprisingly, AZD5305 only worked on defective cells other than normal cells. This preclinical study strongly supports the hypothesis that only inhibiting PARP1 might reduce adverse events without compromising with therapeutical efficacy.55 The ongoing phase I/IIa PETRA trial (NCT04644068) has enrolled 61 patients with advanced HER2-negative breast, ovarian, prostate, or pancreatic cancer who had germline or somatic BRCA1/2, PALB2, or RAD51C/D loss-of-function mutations. According to the preliminary dose-limiting toxicity (DLT) assessment, AZD5305 showed promising clinical activity and safety in those patients.56 Its therapeutic effect in clinical patients and more specific indications for patient selection need further research in the near future.
Mechanism of PARPi resistance
Although PARPi could bring durable survival benefits for selected patients in clinical trials and in the real world, innate and acquired resistance to PARPi has dampened the initial enthusiasm. The underlying mechanisms by which tumor cells evade therapeutic intervention and acquire drug resistance are very complex (Fig. 3). Moreover, the drug resistance mechanisms in each tumor may be the product of several drug resistance mechanisms rather than a single independent mechanism.57 Summaries of the potential drug resistance mechanism studies of different PARP inhibitors are listed in Table 2.
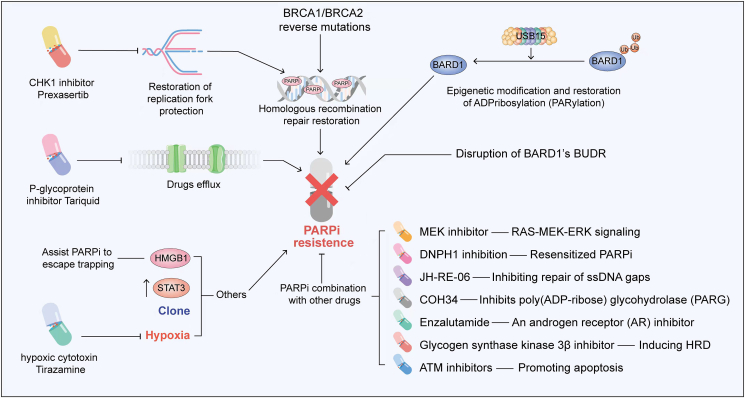
Figure 3.
Multiple mechanisms and potential solutions of PARPi resistance. The mechanisms of PARPi resistance are complex and mainly have the following ways: (i) the increase of drug efflux; (ii) genomic reversal of BRCA1/2; (iii) restoration of replication fork protection; (iv) any strengthening of the homologous recombination repair process; (v) epigenetic modifications; and (vi) other mechanisms, such as high mobility group box 3 (HMGB3), STAT3, clonal selection, and hypoxia. This figure illustrates the main mechanisms of PARPi resistance and the combination strategies that potentially overcome PARPi resistance.
Table 2.
The relevant drug resistance mechanism studies of different PARPi.
| Drug | Mechanism of PARPi resistance | Solution | Reference |
|---|---|---|---|
| Olaparib | Enhancement of drug efflux | p-glycoprotein inhibitor | Rottenberg, 200858 |
| BRCA reversal mutation | Inhibition of DNA end-joining repair pathways | Tobalina, 202165 | |
| Homologous recombination repair restoration | – | Mirman, 202271 | |
| High C/EBPβ expression induces restoration of HR capacity | Targeting C/EBPβ | Tan, 202175 | |
| Restoration of replication fork stability | – | Taglialatela, 201777 | |
| Promotes degradation of stalled replication forks | – | Rondinelli, 201778 | |
| The overexpression of High-mobility group box 3 (HMGB3) increased the insensitivity to PARPi, | Targeted inhibition of HMGB3 | Ma, 202289 | |
| Activates STAT3 | – | Martincuks, 202193 | |
| Clonal evolution | – | Anniina, 202194 | |
| Hypoxia | Eliminating hypoxic tumor cells | Mehibel, 202197 | |
| Rucaparib | BRCA reversal mutation | Improve the detection of mutations | Lin, 201967 |
| Promotes degradation of stalled replication forks | – | Rondinelli, 201778 | |
| Acquired RAD51C promoter methylation loss | – | Ksenija, 202182 | |
| Methylation of BRCA1 copies | quantitative BRCA1 methylation analysis | Kondrashova, 201882 | |
| Niraparib | Acquired RAD51C promoter methylation loss | – | Ksenija, 202183 |
| Clonal evolution | – | Anniina, 202194 | |
| Talazoparib | Homologous recombination repair restoration | – | Waks, 202065 |
| Clonal evolution | – | Anniina, 202194 | |
| Hypoxia | Eliminating hypoxic tumor cells | Mehibel, 202197 |
Drug efflux
Since PARPi needs to enter tumor cells to kill them, an increase in drug efflux is inevitable, affecting the therapeutic effect and eventually leading to drug resistance. The multidrug resistance protein (MDR1) encoded by ATP binding cassette subfamily B member 1 (ABCB1), is involved in the efflux of chemotherapeutic drugs and affects their effectiveness and accumulation in the brain and other tissues. Sven et al found that long-term treatment with olaparib leads to the development of drug resistance due to the up-regulation of ABCB1 encoding p-glycoprotein efflux pumps.58 This resistance to olaparib can be reversed by the addition of p-glycoprotein inhibitor tariquid.58 Therefore, inhibition of ABCB1 may have important clinical implications for patients who are resistant to PARPi.
BRCA1/BRCA2 reverse mutations
Genomic reversal of BRCA1/2 is one of the common molecular mechanisms of PARPi resistance. Patients with defective DNA repair may recover BRCA1 or BRCA2 function through somatic reversion mutation, leading to resistance to PARPi. In several retrospective analyses of next-generation sequencing (NGS) data from 23,375 patients across various common cancer types, the prevalence of multiple pathogenic/likely pathogenic (P/LP) germline mutations in homologous recombination repair (HRR) genes was analyzed.59, 60, 61 The reversion mutations were detected in BRCA1 (3.8%), BRCA2 (3.5%), and PALB2 (2.0%) after platinum-based chemotherapy and/or PARPi resistance.61 The incidence of mutations that restore BRCA1/2 function in ovarian cancer ranges from 0% to 21%, and as high as 40% to 50% in breast cancer.62, 63, 64 A pooled analysis of several studies on reversion mutations revealed mutagenic end-joining DNA repair pathways, particularly those affecting BRCA2, play a key role in the generation of reversion, as indicated by a significant accumulation of microhomology in deletions surrounding DNA sequences leading to reversion events.65 The circulating cell-free DNA (cfDNA) sequencing analysis can help identify BRCA1/2 reversal mutations in breast and ovarian cancer patients.63,66 Lin et al found that in the patients treated with rucaparib, those without BRCA reversal mutations detected in pretreated cfDNA had significantly longer PFS than those with reversal mutations. BRCA reversed mutations detected in cfDNA of platinum-resistant high-grade ovarian carcinoma (HGOC) are associated with the reduced survival benefit of rucaparib therapy.67 Therefore, developing diagnostics for BRCA1/2 reversion mutations could pave the way for overcoming PARPi resistance.
Homologous recombination repair restoration
Any strengthening of the HRR process may give rise to the occurrence of PARPi resistance. 53BP1 is thought to control the fidelity and the choice of DSB repair pathway.68 Waks et al found that in patients with gBRCA1 mutations, TP53BP1 loss or MRE11A amplification had genomic changes that restored HR by increasing DNA terminal excision.64 Shieldin is formed by C20orf196 (also known as SHLD1), FAM35A (SHLD2), CTC-534A2.2 (SHLD3), and REV7, as a downstream effector of 53BP1/RIF1/MAD2L2.69 Those could promote DSBs terminal connection by limiting DSBs resection.70 Recent studies found that the role of 53BP1 and Shieldin proteins in DSB depends on CST–Polα–primase fill-in synthesis and, to some extent, determines the effectiveness of PARPi on BRCA1-deficient cells.71 HR is counteracted by antagonizing BRCA2/RAD51 loading into BRCA1-deficient cells.72,73 Additionally, He et al identified DYNLL1 (dynein light chain 1 protein) as a DNA terminal excision inhibitor that could directly bind to MRE11 and reduce DNA terminal excision activity in vitro. The loss of DYNLL1 enables BRCA1 mutant cells to undergo DNA terminal excision and resume HR. Therefore, the loss and decreased expression of DYNLL1 leads to resistance to platinum drugs and PARPi.74 CCAAT/enhancer binding protein β (C/EBPβ) is an essential transcription factor of the HR pathway. Multiple HR genes (BRCA1, RAD51, BRIT1, and BRIP1) are targeted and up-regulated by C/EBPβ to induce the recovery of HR ability and mediate acquired resistance to PARPi. Deletion of C/EBPβ may be an approach to overcome resistance to PARPi.75
Restoration of replication fork protection
Acquired resistance to PARPi or platinum is in connection with replication fork protection. Chaudhuri et al found that the deletion of the MLL3/4 complex protein PTIP is a key factor to protect BRCA1/2 deficient cells from DNA damage. PTIP deficiency not only restores HR activity at DSBs but also inhibits the recruitment of MRE11 nuclease to the stopped replication fork, which showed in turn the ability to protect the newly formed DNA strand from extensive degradation.76 In BRCA2-deficient tumors without BRCA2 reversion mutations, the acquisition of resistance to PARPi is associated with replication fork protection. Taglialatela et al found that deletion of SMARCAL1, an SNF2 family DNA translocation enzyme, could remodel stalled replication fork and restored its stability, as well as minimize replication stress-induced DNA breaks and chromosomal aberration formation in BRCA1/2 mutated cells.77 It has been found that EZH2 locates at a fork of stasis, where it methylates Lys27 on histone 3 (H3K27me3) to mediate the recruitment of MUS81 nuclease.76 Inhibition of EZH2 in a mouse BRCA2−/− breast cancer model is associated with acquired PARPi resistance. Thus, loss of the EZH2/MUS81 axis function could promote the resistance of BRCA2-deficient cells to PARPi. These findings suggest that low expression of EZH2/MUS81 predicts PARPi resistance and worse outcomes in BRCA2 mutated tumors.78
Epigenetic modification and restoration of ADPribosylation (PARylation)
Epigenetic modifications may also affect sensitivity to PARPi, leading to acquired resistance to PARPi. Some preclinical studies found that silencing BRCA1 or RAD51C genes by methylation leads to HRR defects (HRD).79 The methylation of BRCA1 promoter was first identified in sporadic breast cancer in 1997, and PARPi therapy was subsequently recommended for BRCA1 methylated cancers.80,81 Demethylating agent 5-azacytidine can eliminate PARPi sensitivity in BRCA1-methylated breast cancer cell lines.79 Kondrashova reported that the efficacy of rucaparib was demonstrated in PDX models using 12 high-grade serous ovarian carcinoma (HGSOC) patients.82 Furthermore, if multiple copies of BRCA1 are present in a tumor, all copies must be methylated to develop a PARPi response, and the loss of only one methylated copy of BRCA1 is sufficient to restore HRR DNA repair and induce PARPi resistance. Response to rucaparib prompted that heterozygous methylation is also associated with PARPi resistance. Loss of methylation in the BRCA1 promoter driven by prior chemotherapy increases the likelihood of acquired resistance to PARPi.82 Homozygous RAD51C methylation is also a potential predictive biomarker of sensitivity to PARPi. Collectively, a single non-methylated copy of the gene is sufficient for the development of resistance.83
Others
High mobility group box 3 (HMGB3) is highly expressed in stem cells and cancer cells and is rarely activated in normal adult tissue. This suggests that HMGB3 is a promising therapeutic target.84,85 HMGB3 is associated with radioresistance in cervical cancer,86 and tamoxifen resistance in breast cancer.87 A recent study showed that targeting HMGB3 consumption by inhibiting ATR/CHK1/p-CHK1 DNA damage signaling pathway sensitizes chemotherapy-resistant ovarian cancer cells to cisplatin.88 Ma et al found that HMGB3 was abnormally overexpressed in HGSOC tissues, and the high level of HMGB3 was associated with a shorter OS and a higher risk of resistance to PARPi. PARP1 was identified as a new interaction partner of HMGB3, and the loss of HMGB3 resulted in the trapping of PARP1 at DNA damage sites. Therefore, HMGB3 interaction with PARP1 facilitates its escape from trapping and promotes PARPi resistance.89
Signal transducer and activator of transcription 3 (STAT3), known as a transcription factor,90 can play a role in promoting tumor by inhibiting anti-tumor immune response.91 It has been shown that siRNA-mediated PARP1 or a drug that inhibits PARP1 could lead to increased STAT3 phosphorylation in ovarian cancer cell lines.92 Whether PARPi therapy promotes drug resistance by activating STAT3 in ovarian cancer patients has become a question worth exploring. Martincuks et al found that STAT3 activity was significantly enhanced in tumor cells, tumor-associated immune cells, and fibroblasts after PARPi treatment. This increase in activity leads to PARPi resistance and immunosuppression. Elimination of STAT3 may inhibit the growth of PARPi-resistant ovarian tumor cells and restore the sensitivity to PARPi.93
Cloning may also play a role in drug resistance. A recent study94 used CRISPR/Cas9 technology to obtain TP53 and BRCA1 knockout epithelial cell lines. Seven single-cell clones with acquired resistance after PARPi treatment were obtained. These clones presented increased levels of genomic instability and lower mutational burden with unique transcriptional and mutational profiles compared with PARPi-sensitive cell lines. Clonal analysis showed that acquired PARPi resistance variants were produced by clonal selection. In an image-based drug sensitivity analysis, these clones showed a heterogeneous response pattern based on diverse resistance molecular mechanisms, thus there is an urgent to identify vulnerabilities to the selected agents.94
Oxygen levels inside the tumor are uneven, reaching as high as 2% in some areas and less than 0.01% in others. Hypoxia, a property not found in normal cells, could be a target for cancer treatment.95 Hypoxia, as a hallmark feature of the tumor microenvironment, could induce resistance to chemotherapy.96 Moderate hypoxia (oxygen <2%) promotes the resistance of HR-proficient and/or deficient cancer cells to PARPi in a HIF-independent pathway.97 However, some studies have shown that the anoxic conditions with oxygen content less than 0.2% can increase the sensitivity of HR-proficient tumors to PARPi with synthetic lethal effects.98 Theoretically, the reduction of ROS-induced DNA damage was responsible for the observed resistance. Remarkably, it has been shown that the hypoxic cytotoxin tirapazamine, targeting hypoxic tumor cells, could enhance the efficacy of PARPi therapy.97
To sum up, PARPi has a variety of resistance mechanisms. BRCA1/BRCA2 reverse mutations,60 epigenetic modification and restoration of ADPribosylation (PARylation),82 restoration of homologous recombination repair75 or replication fork protection76 could be found in ovarian cancer. Drug efflux,58 BRCA1/BRCA2 reverse mutations,59 epigenetic modification and restoration of PARylation79 were found in breast cancer. However, among those different resistance mechanisms, only HR reverse mutations have been found in patients in clinical trials. Therefore, further research is needed to provide evidence of all the different resistance mechanisms in clinical trials.
Solutions to PARPi resistance
Overcoming PARPi resistance and identifying predictive biomarkers for PARPi response have been investigated in recent years and will have a great impact in the era of precision medicine.
Identifying novel predictive biomarkers
Several studies have revealed that RAD51 has the potential to be a novel predictive biomarker.38,99, 100, 101 The detection of RAD51 foci using immunohistochemistry was associated with the functional status of BRCA1/2 shown in genomic data, as well as the response to DNA damage treatment, supporting the formation of these foci as a clinically useful biomarker.57,65 Down-regulation of RAD51 using shRNA sensitized cancer stem cells (CSCs) to PARPi and inhibited tumor growth in triple-negative breast cancer.102 Bermejo et al analyzed the predictive function of RAD51 score and HR defect in breast cancer PDXs, reporting a clear distinction in RAD51 between PARPi-sensitive and -resistant cells. The RAD51 score outperformed gene sequencing in terms of detecting PARPi sensitivity and resistance.100,101 The above studies indicate that RAD51 is a valuable biomarker for predicting PARPi resistance.
The identification of other RAD51-like biomarkers will expand the use of PARPi in more clinical applications and benefit more patients. After reviewing the molecular profiles of 52,426 tumor samples from 21 cancer types, Heeke AL et al identified the frequencies of pathogenic mutations in HR-DDR genes (ARID1A, ATM, ATRX, BAP1, BARD1, BLM, BRCA1/2, BRIP1, CHEK1/2, FANCA/C/D2/E/F/G/L, MRE11A, NBN, PALB2, RAD50, RAD51, RAD51B, and WRN). The frequencies of HR mutations were 15.6% in triple-negative breast cancers, 18.1% in melanoma, 15.4% in pancreatic cancer, and 15.0% in CRC among others. Regarding the pathogenic mutation frequency of the HR pathway genes, ARID1A (7.2%) was the most mutated gene in HR pathway, followed by other 14 genes including BRCA2 (3.0%), BRCA1 (2.8%), ATM (1.3%), ATRX (1.3%), and CHEK2 (1.3%).103 The proportion of mutation frequency of these 15 genes is demonstrated in Figure 4. Recently, Ipsen et al identified six candidate genes with the utility of genome-wide CRISPR-Cas9. Multiple knockout populations/clones of each of the six genes were then generated in C4 and/or LNCaP CRPC cells, confirming that the deletion of PARP1, ARH3, YWHAE, or UBR5 caused resistance to olaparib. Drug resistance caused by the knockout of PARP1 and ARH3 may be related to decreased autophagy, but further research is needed.104 Peng et al found that the deubiquitylating enzyme USP15 regulates homologous recombination repair and attenuates the response to PARPi in cancer cells. USP15 is recruited to DSBs and deubiquitinates the BRCT domain of BRCA1-associated RING domain protein 1 (BARD1) to promote BRCA1/BARD1 retention and function in DSBs. Consequently, overexpression of USP15 will largely contribute to the occurrence of drug resistance.105 In the near future, advanced sequencing techniques can be used to select PARPi-sensitive patients and reduce the occurrence of drug resistance.
Figure 4.
The mutation frequencies of 15 HR-DDR genes. The HR-DDR genes shown here are ARID1A, ATM, ATRX, BAP1, BARD1, BLM, BRCA1/2, BRIP1, CHEK1/2, FANCA/C/D2/E/F/G/L, MRE11A, NBN, PALB2, RAD50, RAD51, RAD51B, and WRN.
Combination with other targeted therapies
Numerous studies have investigated how to overcome PARPi resistance pathways based on molecular mechanisms. Some studies have made breakthrough progress in drug combination. The combination of PARPi and other targeted drugs can make patients re-sensitive to PARPi, or improve the efficacy to achieve synthetic lethality.
First, unexpected synergistic cytotoxic effects were achieved by the addition of mitogen-activated protein kinase (MAPK) kinase (MEK) inhibitors to PARPi. Sun et al found that RAS-MEK-ERK signaling in cancer cells is up-regulated in multiple RAS-mutated tumor models in vivo and in vitro.106 RAS pathways are also activated in PARPi-resistant cells. The efficacy of PARPi combined with MEK inhibitor was independent of the mutational status of BRCA1/2 and TP53, suggesting that synergies may be expandable.106 Moreover, MEK inhibitors can reverse resistance to PARPi therapy.107 These results indicate that the addition of MEK inhibitor to PARPi not only improves the efficacy but also overcomes drug resistance.
Second, from the perspective of the structure and mechanism of PARPi action, the resistance to PARPi can be reversed by inhibiting P protein or exposing ssDNA gap or dePARylation. The reduction in intracellular P-glycoprotein concentration by efflux of PARPi leads to resistance to PARPi. This re-sensitization to PARPi can be found by combination therapy with the p-glycoprotein inhibitor tariquidar.58 In addition, the ssDNA gap is also an important therapeutic target for drug resistance. When conducting functional genetic screening in PARPi-resistant cells and organoids through 53BP1 deletion, Paes Dias et al found that deletion of nuclear DNA ligase III (LIG3) could enhance PARPi toxicity in BRCA1-deficient cells. It was found that LIG3 deletion promoted the formation of MRE11-mediated post-replication ssDNA gaps in BRCA1-deficient and BRCA1/53BP1 double-deficient cells, and then increase the exposure to PARPi and could reverse PARPi resistance in BRCA1/53BP1 double-deficient cells.108 JH-RE-06 has a potentially toxic effect on PARPi-resistant BRCA1 mutant cells by inhibiting the repair of ssDNA gaps. Furthermore, it displays additive toxicity with crosslinking agents or PARPi.109 With the utility of COH34, a small molecule targeting poly (ADP-ribose) glycohydrolase (PARG), Chen et al identified its novel role of dePARylation in DNA repair. COH34 binds to the CAT domain of PARG, a major dePARylation enzyme, and then could prolong PARylation at the site of DNA damage and trap DNA repair factors. COH34 was shown not only to have a lethal effect on cancer cells with DNA repair deficiency but also to effectively kill PARPi-resistant cancer cells.110 Prexasertib, a checkpoint kinase 1 (CHK1) inhibitor, can lead to replication catastrophe in PARPi-resistant HGSOC PDX models and cell lines. The combination of olaparib with prexasertib could not only significantly slow the tumor growth in olaparib-resistant models, but also further enhance the magnitude and persistence of response in olaparib-sensitive models. Moreover, prexasertib reverses restored HR and replication fork stability, acting synergistically with PARPi.111
Third, combination with other targeted agents could enhance the anti-tumor effect of PARPi. Two recent studies have shown that pyruvate kinase M2 (PKM2) inhibitor and ALK kinase inhibitor ceritinib could induce DNA damage in ovarian cancer cells and improve the response to olaparib, respectively.112,113 Gabbasov et al revealed heat shock protein 90 (HSP90) regulates the maturation and stability of a key protein required for DDR. It also indicates that the addition of ganetespib, a unique small molecule HSP90 inhibitor, could effectively disrupt key DDR pathway proteins and make ovarian cancer cells without “BRCAness” respond to talazoparib.113,114 In addition, combination therapy also works in breast cancer. Wang et al demonstrated that the limited response of BRCA1-deficient breast tumors to PARPi is mainly due to M2-like pro-tumor macrophages, which inhibits CD8+ T cells in the immune system and impedes PARPi-triggered tumor cell DNA damage as well. The addition of exogenous STING agonists can transform tumor-associated macrophages (TAMs) from M2-like into an M1-like anti-tumor state. Moreover, systemic administration of STING agonist combined with PARPi enhances anti-tumor immunity regardless of STING expression in tumor cells and has shown significant therapeutic effect in BRCA1-deficient breast cancer mouse models.115 Li et al found that enzalutamide, an inhibitor of androgen receptor, inhibits the expression of several genes associated with HR in CRPC cell lines, even though it is generally ineffective in CRPC patients. Therefore, the combination of olaparib with enzalutamide could promote cell death in CRPC cells.116 Blocking nucleotide salvage factor DNPH1 combined with the treatment with hmdU (5-hydroxymethyl-deoxyuridine) could overcome the resistance of PARPi in BRCA1-deficient cells. Targeting DNPH1 provides a promising strategy for the hypersensitization of BRCA-deficient cancers to PARPi therapy.117 In addition, combining PARPi with molecules that target cell cycle checkpoints is also a synergistic approach. The DNA replication stress checkpoint (ATR-CHK1-WEE1) showed promising results among cell cycle checkpoints.118 When combined with olaparib, the WEE1 inhibitor provides a unique therapeutic strategy to overcome PARPi resistance by targeting replication stress response.119 Kim et al found that the acquired resistance to PARPi was often accompanied by increased ATR-CHK1 activity and ATR inhibitor (ATRi) sensitivity. In a clinically relevant acquired PARPi-resistant PDXs model, a significant increase in survival rate was observed with a durable complete therapeutic response to the combination of PARPi and ATRi.120 More robust clinical trial evidence in the future is needed to select the optimal treatment strategy for patients.
The combination of radiotherapy and immunotherapy
Radiation causes DNA damage in cells, including DSBs, SSBs, and interstrand cross-linking, which disrupts DNA replication and the following transcription and ultimately leads to cell death.121 PARP1 inhibitor could enhance ionizing radiation (IR)-induced cytotoxicity by inhibiting NF-κB activation.122 Also, PARP2 depletion demonstrated higher sensitivity to the cell-killing effects of IR in vitro.123 Preliminary data showed that PARPi or PARP depletion has a radiosensitization effect to enhance the efficacy of radiotherapy in ovarian cancer, breast cancer, cholangiocarcinoma, and soft tissue sarcoma.124, 125, 126, 127 The addition of DNA methyltransferase inhibitors (DNMTis) to PARPi could further improve the response in NSCLC cells to IR in vitro and in vivo.126 Since nitric oxide enhances HRD by inhibiting BRCA1 expression under oxidative stress, nitric oxide-donor combined with PARPi provides a new approach for sensitization to IR.128 These preclinical studies suggested a synergistic effect when combining radiation with PARPi therapy, indicating promising investigations in clinical trials. Several studies have explored how to maximize the benefit of combination therapy in vitro. The low dosage of olaparib is associated with higher sensitivity to the combination therapy, particularly in cells with homologous recombination-impaired status.129 In HR-proficient tumor types, p53 status was a candidate predictive biomarker, which could be used to select patients who might benefit more from the combination therapy.130
Immuno-oncology (IO) therapy activates the ability of the immune system to eliminate cancer cells rather than targeting the tumor directly.131 PARPi could transform “cold” tumors into “hot” tumors to improve the response to immunotherapy. PARPi not only increases tumor mutation burden to generate more neoantigens by inhibiting DNA damage repair, but it also induces the expression of PD-L1.132 The rationale for the combination of PARPi and immunotherapy has led to numerous subsequent clinical trials.133 Pembrolizumab, a PD-1 inhibitor that can block the interaction between PD-1 and its ligands PD-L1/PD-L2, received FDA approval for TNBC in neoadjuvant and metastatic settings. The addition of pembrolizumab to niraparib showed promising anti-tumor activity in advanced TNBC,134 refractory platinum-resistant ovarian cancer,135 and mCRPC.136 There are ongoing studies on the combination of PARPi and anti-PD1/PD-L1 agents in patients with advanced solid cancers.137,138 Encouraging results from these pilot studies revealed the synergistic effect of the combination, warranting further investigation. Also, it should be noted that a subset of patients are not responders, suggesting it is urgent to identify the predictive biomarkers to select patients for combination therapy. Based on the retrospective exploratory research in TOPACIO trial,135 potential predictive biomarkers for the efficacy of the niraparib/pembrolizumab combination are mutational signature 3 and interferon signaling in the CD8+ T cells in the tumor microenvironment, in addition to HR or BRCA mutation status.139 Therefore, predictive biomarkers should be the primary focus of future studies to prospectively identify the ones who would benefit from combination therapy.
Conclusion
Despite the great progress in using PARPi in clinical treatment for HR-deficient tumors, the emerging drug resistance has dampened the initial enthusiasm. Deeply understanding the mechanisms of resistance to PARPi and developing novel combination therapies to overcome drug resistance are essential to optimize the use of PARPi and benefit more patients.
Conflict of interests
The authors declare no conflict of interests.
Footnotes
Peer review under responsibility of Chongqing Medical University.
Contributor Information
Huadong Pei, Email: huadong.pei@georgetown.edu.
Juanjuan Li, Email: juanjuan.li@whu.edu.cn.
References
- 1.Lindahl T., Satoh M.S., Poirier G.G., et al. Post-translational modification of poly(ADP-ribose) polymerase induced by DNA strand breaks. Trends Biochem Sci. 1995;20(10):405–411. doi: 10.1016/s0968-0004(00)89089-1. [DOI] [PubMed] [Google Scholar]
- 2.Gibson B.A., Kraus W.L. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol. 2012;13(7):411–424. doi: 10.1038/nrm3376. [DOI] [PubMed] [Google Scholar]
- 3.Amé J.C., Spenlehauer C., de Murcia G. The PARP superfamily. Bioessays. 2004;26(8):882–893. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- 4.Hassa P.O., Hottiger M.O. The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front Biosci. 2008;13:3046–3082. doi: 10.2741/2909. [DOI] [PubMed] [Google Scholar]
- 5.Langelier M.F., Planck J.L., Roy S., et al. Structural basis for DNA damage-dependent poly(ADP-ribosyl)ation by human PARP-1. Science. 2012;336(6082):728–732. doi: 10.1126/science.1216338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Langelier M.F., Pascal J.M. PARP-1 mechanism for coupling DNA damage detection to poly(ADP-ribose) synthesis. Curr Opin Struct Biol. 2013;23(1):134–143. doi: 10.1016/j.sbi.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caldecott K.W. Mammalian DNA base excision repair: dancing in the moonlight. DNA Repair. 2020;93 doi: 10.1016/j.dnarep.2020.102921. [DOI] [PubMed] [Google Scholar]
- 8.Beard W.A., Horton J.K., Prasad R., et al. Eukaryotic base excision repair: new approaches shine light on mechanism. Annu Rev Biochem. 2019;88:137–162. doi: 10.1146/annurev-biochem-013118-111315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Demin A.A., Hirota K., Tsuda M., et al. XRCC1 prevents toxic PARP1 trapping during DNA base excision repair. Mol Cell. 2021;81(14):3018–3030.e5. doi: 10.1016/j.molcel.2021.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao Y., Li C., Wei L., et al. SSRP1 cooperates with PARP and XRCC1 to facilitate single-strand DNA break repair by chromatin priming. Cancer Res. 2017;77(10):2674–2685. doi: 10.1158/0008-5472.CAN-16-3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Q., Ma K., Liu X., et al. Truncated PARP1 mediates ADP-ribosylation of RNA polymerase III for apoptosis. Cell Discov. 2022;8:3. doi: 10.1038/s41421-021-00355-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibson B.A., Zhang Y., Jiang H., et al. Chemical genetic discovery of PARP targets reveals a role for PARP-1 in transcription elongation. Science. 2016;353(6294):45–50. doi: 10.1126/science.aaf7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sung P., Klein H. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat Rev Mol Cell Biol. 2006;7(10):739–750. doi: 10.1038/nrm2008. [DOI] [PubMed] [Google Scholar]
- 14.Yap T.A., Sandhu S.K., Carden C.P., et al. Poly(ADP-ribose) polymerase (PARP) inhibitors: exploiting a synthetic lethal strategy in the clinic. CA Cancer J Clin. 2011;61(1):31–49. doi: 10.3322/caac.20095. [DOI] [PubMed] [Google Scholar]
- 15.Ang W.H., Myint M., Lippard S.J. Transcription inhibition by platinum-DNA cross-links in live mammalian cells. J Am Chem Soc. 2010;132(21):7429–7435. doi: 10.1021/ja101495v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fong P.C., Yap T.A., Boss D.S., et al. Poly(ADP)-ribose polymerase inhibition: frequent durable responses in BRCA carrier ovarian cancer correlating with platinum-free interval. J Clin Oncol. 2010;28(15):2512–2519. doi: 10.1200/JCO.2009.26.9589. [DOI] [PubMed] [Google Scholar]
- 17.Ledermann J., Harter P., Gourley C., et al. Olaparib maintenance therapy in patients with platinum-sensitive relapsed serous ovarian cancer: a preplanned retrospective analysis of outcomes by BRCA status in a randomised phase 2 trial. Lancet Oncol. 2014;15(8):852–861. doi: 10.1016/S1470-2045(14)70228-1. [DOI] [PubMed] [Google Scholar]
- 18.Pujade-Lauraine E., Ledermann J.A., Selle F., et al. Olaparib tablets as maintenance therapy in patients with platinum-sensitive, relapsed ovarian cancer and a BRCA1/2 mutation (SOLO2/ENGOT-Ov21):a double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol. 2017;18(9):1274–1284. doi: 10.1016/S1470-2045(17)30469-2. [DOI] [PubMed] [Google Scholar]
- 19.Citarelli M., Teotia S., Lamb R.S. Evolutionary history of the poly(ADP-ribose) polymerase gene family in eukaryotes. BMC Evol Biol. 2010;10:308. doi: 10.1186/1471-2148-10-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gagné J.P., Isabelle M., Lo K.S., et al. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res. 2008;36(22):6959–6976. doi: 10.1093/nar/gkn771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.El-Khamisy S.F., Masutani M., Suzuki H., et al. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 2003;31(19):5526–5533. doi: 10.1093/nar/gkg761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lord C.J., Ashworth A. PARP inhibitors: synthetic lethality in the clinic. Science. 2017;355(6330):1152–1158. doi: 10.1126/science.aam7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bell N.A.W., Haynes P.J., Brunner K., et al. Single-molecule measurements reveal that PARP1 condenses DNA by loop stabilization. Sci Adv. 2021;7(33) doi: 10.1126/sciadv.abf3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langelier M.F., Eisemann T., Riccio A.A., et al. PARP family enzymes: regulation and catalysis of the poly(ADP-ribose) posttranslational modification. Curr Opin Struct Biol. 2018;53:187–198. doi: 10.1016/j.sbi.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zandarashvili L., Langelier M.F., Velagapudi U.K., et al. Structural basis for allosteric PARP-1 retention on DNA breaks. Science. 2020;368(6486) doi: 10.1126/science.aax6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Helleday T. The underlying mechanism for the PARP and BRCA synthetic lethality: clearing up the misunderstandings. Mol Oncol. 2011;5(4):387–393. doi: 10.1016/j.molonc.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saleh-Gohari N., Bryant H.E., Schultz N., et al. Spontaneous homologous recombination is induced by collapsed replication forks that are caused by endogenous DNA single-strand breaks. Mol Cell Biol. 2005;25(16):7158–7169. doi: 10.1128/MCB.25.16.7158-7169.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krastev D.B., Li S., Sun Y., et al. The ubiquitin-dependent ATPase p97 removes cytotoxic trapped PARP1 from chromatin. Nat Cell Biol. 2022;24(1):62–73. doi: 10.1038/s41556-021-00807-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cong K., Peng M., Kousholt A.N., et al. Replication gaps are a key determinant of PARP inhibitor synthetic lethality with BRCA deficiency. Mol Cell. 2021;81(15):3128–3144.e7. doi: 10.1016/j.molcel.2021.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saha L.K., Murai Y., Saha S., et al. Replication-dependent cytotoxicity and Spartan-mediated repair of trapped PARP1-DNA complexes. Nucleic Acids Res. 2021;49(18):10493–10506. doi: 10.1093/nar/gkab777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mateo J., Lord C.J., Serra V., et al. A decade of clinical development of PARP inhibitors in perspective. Ann Oncol. 2019;30(9):1437–1447. doi: 10.1093/annonc/mdz192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dias M.P., Moser S.C., Ganesan S., et al. Understanding and overcoming resistance to PARP inhibitors in cancer therapy. Nat Rev Clin Oncol. 2021;18(12):773–791. doi: 10.1038/s41571-021-00532-x. [DOI] [PubMed] [Google Scholar]
- 33.Fong P.C., Boss D.S., Yap T.A., et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361(2):123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 34.Banerjee S., Moore K.N., Colombo N., et al. Maintenance olaparib for patients with newly diagnosed advanced ovarian cancer and a BRCA mutation (SOLO1/GOG 3004):5-year follow-up of a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2021;22(12):1721–1731. doi: 10.1016/S1470-2045(21)00531-3. [DOI] [PubMed] [Google Scholar]
- 35.Frenel J.S., Kim J.W., Aryal N., et al. Efficacy of subsequent chemotherapy for patients with BRCA1/2-mutated recurrent epithelial ovarian cancer progressing on olaparib versus placebo maintenance: post-hoc analyses of the SOLO2/ENGOT Ov-21 trial. Ann Oncol. 2022;33(10):1021–1028. doi: 10.1016/j.annonc.2022.06.011. [DOI] [PubMed] [Google Scholar]
- 36.Robinson A.G., Izard J.P., Vera-Badillo F.E. Treatment and patient selection for patients with metastatic castration-resistant prostate after progression on docetaxel and abiraterone/enzalutamide: when to play your CARD and when to do your PARP. Eur Urol. 2021;80(2):123–126. doi: 10.1016/j.eururo.2021.03.001. [DOI] [PubMed] [Google Scholar]
- 37.Mateo J., Porta N., Bianchini D., et al. Olaparib in patients with metastatic castration-resistant prostate cancer with DNA repair gene aberrations (TOPARP-B): a multicentre, open-label, randomised, phase 2 trial. Lancet Oncol. 2020;21(1):162–174. doi: 10.1016/S1470-2045(19)30684-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carreira S., Porta N., Arce-Gallego S., et al. Biomarkers associating with PARP inhibitor benefit in prostate cancer in the TOPARP-B trial. Cancer Discov. 2021;11(11):2812–2827. doi: 10.1158/2159-8290.CD-21-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Bono J., Mateo J., Fizazi K., et al. Olaparib for metastatic castration-resistant prostate cancer. N Engl J Med. 2020;382(22):2091–2102. doi: 10.1056/NEJMoa1911440. [DOI] [PubMed] [Google Scholar]
- 40.Mateo J., Carreira S., Sandhu S., et al. DNA-repair defects and olaparib in metastatic prostate cancer. N Engl J Med. 2015;373(18):1697–1708. doi: 10.1056/NEJMoa1506859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Golan T., Hammel P., Reni M., et al. Maintenance olaparib for germline BRCA-mutated metastatic pancreatic cancer. N Engl J Med. 2019;381(4):317–327. doi: 10.1056/NEJMoa1903387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reiss K.A., Mick R., O'Hara M.H., et al. Phase II study of maintenance rucaparib in patients with platinum-sensitive advanced pancreatic cancer and a pathogenic germline or somatic variant in BRCA1, BRCA2, or PALB2. J Clin Oncol. 2021;39(22):2497–2505. doi: 10.1200/JCO.21.00003. [DOI] [PubMed] [Google Scholar]
- 43.Swisher E.M., Kwan T.T., Oza A.M., et al. Molecular and clinical determinants of response and resistance to rucaparib for recurrent ovarian cancer treatment in ARIEL2 (Parts 1 and 2) Nat Commun. 2021;12:2487. doi: 10.1038/s41467-021-22582-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.González-Martín A., Pothuri B., Vergote I., et al. Niraparib in patients with newly diagnosed advanced ovarian cancer. N Engl J Med. 2019;381(25):2391–2402. doi: 10.1056/NEJMoa1910962. [DOI] [PubMed] [Google Scholar]
- 45.Mirza M.R., Monk B.J., Herrstedt J., et al. Niraparib maintenance therapy in platinum-sensitive, recurrent ovarian cancer. N Engl J Med. 2016;375(22):2154–2164. doi: 10.1056/NEJMoa1611310. [DOI] [PubMed] [Google Scholar]
- 46.Ai X., Pan Y., Shi J., et al. Efficacy and safety of niraparib as maintenance treatment in patients with extensive-stage SCLC after first-line chemotherapy: a randomized, double-blind, phase 3 study. J Thorac Oncol. 2021;16(8):1403–1414. doi: 10.1016/j.jtho.2021.04.001. [DOI] [PubMed] [Google Scholar]
- 47.Litton J.K., Rugo H.S., Ettl J., et al. Talazoparib in patients with advanced breast cancer and a germline BRCA mutation. N Engl J Med. 2018;379(8):753–763. doi: 10.1056/NEJMoa1802905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Bono J.S., Mehra N., Scagliotti G.V., et al. Talazoparib monotherapy in metastatic castration-resistant prostate cancer with DNA repair alterations (TALAPRO-1): an open-label, phase 2 trial. Lancet Oncol. 2021;22(9):1250–1264. doi: 10.1016/S1470-2045(21)00376-4. [DOI] [PubMed] [Google Scholar]
- 49.Li N., Bu H., Liu J., et al. An open-label, multicenter, single-arm, phase II study of fluzoparib in patients with germline BRCA1/2 mutation and platinum-sensitive recurrent ovarian cancer. Clin Cancer Res. 2021;27(9):2452–2458. doi: 10.1158/1078-0432.CCR-20-3546. [DOI] [PubMed] [Google Scholar]
- 50.Li N., Zhang Y., Wang J., et al. Fuzuloparib maintenance therapy in patients with platinum-sensitive, recurrent ovarian carcinoma (FZOCUS-2): a multicenter, randomized, double-blind, placebo-controlled, phase III trial. J Clin Oncol. 2022;40(22):2436–2446. doi: 10.1200/JCO.21.01511. [DOI] [PubMed] [Google Scholar]
- 51.Murai J., Huang S.Y.N., Das B.B., et al. Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res. 2012;72(21):5588–5599. doi: 10.1158/0008-5472.CAN-12-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ronson G.E., Piberger A.L., Higgs M.R., et al. PARP1 and PARP2 stabilise replication forks at base excision repair intermediates through Fbh1-dependent Rad51 regulation. Nat Commun. 2018;9:746. doi: 10.1038/s41467-018-03159-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Papeo G., Posteri H., Borghi D., et al. Discovery of 2-[1-(4,4-difluorocyclohexyl)piperidin-4-yl]-6-fluoro-3-oxo-2,3-dihydro-1H-isoindole-4-carboxamide (NMS-P118): a potent, orally available, and highly selective PARP-1 inhibitor for cancer therapy. J Med Chem. 2015;58(17):6875–6898. doi: 10.1021/acs.jmedchem.5b00680. [DOI] [PubMed] [Google Scholar]
- 54.Johannes J.W., Balazs A., Barratt D., et al. Discovery of 5-{4-[(7-ethyl-6-oxo-5,6-dihydro-1,5-naphthyridin-3-yl)methyl]piperazin-1-yl}-N-methylpyridine-2-carboxamide (AZD5305): a PARP1-DNA trapper with high selectivity for PARP1 over PARP2 and other PARPs. J Med Chem. 2021;64(19):14498–14512. doi: 10.1021/acs.jmedchem.1c01012. [DOI] [PubMed] [Google Scholar]
- 55.Illuzzi G., Staniszewska A.D., Gill S.J., et al. Preclinical characterization of AZD5305, A next-generation, highly selective PARP1 inhibitor and trapper. Clin Cancer Res. 2022;28(21):4724–4736. doi: 10.1158/1078-0432.CCR-22-0301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yap T.A., Im S., Schram A.M., et al. Presented at American Association for Cancer Research Annual Meeting. 2022. PETRA: First in class, first in human trial of the next generation PARP1-selective inhibitor AZD5305 in patients (pts) with BRCA1/2, PALB2 or RAD51C/D mutations. [Google Scholar]
- 57.Cruz C., Castroviejo-Bermejo M., Gutiérrez-Enríquez S., et al. RAD51 foci as a functional biomarker of homologous recombination repair and PARP inhibitor resistance in germline BRCA-mutated breast cancer. Ann Oncol. 2018;29(5):1203–1210. doi: 10.1093/annonc/mdy099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rottenberg S., Jaspers J.E., Kersbergen A., et al. High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs. Proc Natl Acad Sci U S A. 2008;105(44):17079–17084. doi: 10.1073/pnas.0806092105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Heeke A.L., Xiu J., Elliott A., et al. Actionable co-alterations in breast tumors with pathogenic mutations in the homologous recombination DNA damage repair pathway. Breast Cancer Res Treat. 2020;184(2):265–275. doi: 10.1007/s10549-020-05849-2. [DOI] [PubMed] [Google Scholar]
- 60.Konstantinopoulos P.A., Ceccaldi R., Shapiro G.I., et al. Homologous recombination deficiency: exploiting the fundamental vulnerability of ovarian cancer. Cancer Discov. 2015;5(11):1137–1154. doi: 10.1158/2159-8290.CD-15-0714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zong H., Zhang J., Xu Z., et al. Comprehensive analysis of somatic reversion mutations in homologous recombination repair (HRR) genes in A large cohort of Chinese pan-cancer patients. J Cancer. 2022;13(4):1119–1129. doi: 10.7150/jca.65650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ang J.E., Gourley C., Powell C.B., et al. Efficacy of chemotherapy in BRCA1/2 mutation carrier ovarian cancer in the setting of PARP inhibitor resistance: a multi-institutional study. Clin Cancer Res. 2013;19(19):5485–5493. doi: 10.1158/1078-0432.CCR-13-1262. [DOI] [PubMed] [Google Scholar]
- 63.Weigelt B., Comino-Méndez I., de Bruijn I., et al. Diverse BRCA1 and BRCA2 reversion mutations in circulating cell-free DNA of therapy-resistant breast or ovarian cancer. Clin Cancer Res. 2017;23(21):6708–6720. doi: 10.1158/1078-0432.CCR-17-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Waks A.G., Cohen O., Kochupurakkal B., et al. Reversion and non-reversion mechanisms of resistance to PARP inhibitor or platinum chemotherapy in BRCA1/2-mutant metastatic breast cancer. Ann Oncol. 2020;31(5):590–598. doi: 10.1016/j.annonc.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tobalina L., Armenia J., Irving E., et al. A meta-analysis of reversion mutations in BRCA genes identifies signatures of DNA end-joining repair mechanisms driving therapy resistance. Ann Oncol. 2021;32(1):103–112. doi: 10.1016/j.annonc.2020.10.470. [DOI] [PubMed] [Google Scholar]
- 66.Quigley D., Alumkal J.J., Wyatt A.W., et al. Analysis of circulating cell-free DNA identifies multiclonal heterogeneity of BRCA2 reversion mutations associated with resistance to PARP inhibitors. Cancer Discov. 2017;7(9):999–1005. doi: 10.1158/2159-8290.CD-17-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lin K.K., Harrell M.I., Oza A.M., et al. BRCA reversion mutations in circulating tumor DNA predict primary and acquired resistance to the PARP inhibitor rucaparib in high-grade ovarian carcinoma. Cancer Discov. 2019;9(2):210–219. doi: 10.1158/2159-8290.CD-18-0715. [DOI] [PubMed] [Google Scholar]
- 68.Mirman Z., de Lange T. 53BP1:a DSB escort. Genes Dev. 2020;34(1–2):7–23. doi: 10.1101/gad.333237.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Setiaputra D., Durocher D. Shieldin - the protector of DNA ends. EMBO Rep. 2019;20(5) doi: 10.15252/embr.201847560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Panier S., Boulton S.J. Double-strand break repair: 53BP1 comes into focus. Nat Rev Mol Cell Biol. 2014;15(1):7–18. doi: 10.1038/nrm3719. [DOI] [PubMed] [Google Scholar]
- 71.Mirman Z., Sasi N.K., King A., et al. 53BP1-shieldin-dependent DSB processing in BRCA1-deficient cells requires CST-Polα-primase fill-in synthesis. Nat Cell Biol. 2022;24(1):51–61. doi: 10.1038/s41556-021-00812-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Noordermeer S.M., Adam S., Setiaputra D., et al. The shieldin complex mediates 53BP1-dependent DNA repair. Nature. 2018;560(7716):117–121. doi: 10.1038/s41586-018-0340-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dev H., Chiang T.W.W., Lescale C., et al. Shieldin complex promotes DNA end-joining and counters homologous recombination in BRCA1-null cells. Nat Cell Biol. 2018;20(8):954–965. doi: 10.1038/s41556-018-0140-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.He Y.J., Meghani K., Caron M.C., et al. DYNLL1 binds to MRE11 to limit DNA end resection in BRCA1-deficient cells. Nature. 2018;563(7732):522–526. doi: 10.1038/s41586-018-0670-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tan J., Zheng X., Li M., et al. C/EBPβ promotes poly(ADP-ribose) polymerase inhibitor resistance by enhancing homologous recombination repair in high-grade serous ovarian cancer. Oncogene. 2021;40(22):3845–3858. doi: 10.1038/s41388-021-01788-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chaudhuri A.R., Callen E., Ding X., et al. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature. 2016;535(7612):382–387. doi: 10.1038/nature18325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Taglialatela A., Alvarez S., Leuzzi G., et al. Restoration of replication fork stability in BRCA1- and BRCA2-deficient cells by inactivation of SNF2-family fork remodelers. Mol Cell. 2017;68(2):414–430.e8. doi: 10.1016/j.molcel.2017.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rondinelli B., Gogola E., Yücel H., et al. EZH2 promotes degradation of stalled replication forks by recruiting MUS81 through histone H3 trimethylation. Nat Cell Biol. 2017;19(11):1371–1378. doi: 10.1038/ncb3626. [DOI] [PubMed] [Google Scholar]
- 79.Veeck J., Ropero S., Setien F., et al. BRCA1 CpG island hypermethylation predicts sensitivity to poly(adenosine diphosphate)-ribose polymerase inhibitors. J Clin Oncol. 2010;28(29):e563–e564. doi: 10.1200/JCO.2010.30.1010. author reply e565-6. [DOI] [PubMed] [Google Scholar]
- 80.Dobrovic A., Simpfendorfer D. Methylation of the BRCA1 gene in sporadic breast cancer. Cancer Res. 1997;57(16):3347–3350. [PubMed] [Google Scholar]
- 81.Ashworth A. A synthetic lethal therapeutic approach: poly(ADP) ribose polymerase inhibitors for the treatment of cancers deficient in DNA double-strand break repair. J Clin Oncol. 2008;26(22):3785–3790. doi: 10.1200/JCO.2008.16.0812. [DOI] [PubMed] [Google Scholar]
- 82.Kondrashova O., Topp M., Nesic K., et al. Methylation of all BRCA1 copies predicts response to the PARP inhibitor rucaparib in ovarian carcinoma. Nat Commun. 2018;9:3970. doi: 10.1038/s41467-018-05564-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nesic K., Kondrashova O., Hurley R.M., et al. Acquired RAD51C promoter methylation loss causes PARP inhibitor resistance in high-grade serous ovarian carcinoma. Cancer Res. 2021;81(18):4709–4722. doi: 10.1158/0008-5472.CAN-21-0774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gao J., Aksoy B.A., Dogrusoz U., et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wilke K., Wiemann S., Gaul R., et al. Isolation of human and mouse HMG2a cDNAs: evidence for an HMG2a-specific 3' untranslated region. Gene. 1997;198(1–2):269–274. doi: 10.1016/s0378-1119(97)00324-7. [DOI] [PubMed] [Google Scholar]
- 86.Li Z., Zhang Y., Sui S., et al. Targeting HMGB3/hTERT axis for radioresistance in cervical cancer. J Exp Clin Cancer Res. 2020;39:243. doi: 10.1186/s13046-020-01737-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li X., Wu Y., Liu A., et al. miR-27b is epigenetically downregulated in tamoxifen resistant breast cancer cells due to promoter methylation and regulates tamoxifen sensitivity by targeting HMGB3. Biochem Biophys Res Commun. 2016;477(4):768–773. doi: 10.1016/j.bbrc.2016.06.133. [DOI] [PubMed] [Google Scholar]
- 88.Mukherjee A., Huynh V., Gaines K., et al. Targeting the high-mobility group box 3 protein sensitizes chemoresistant ovarian cancer cells to cisplatin. Cancer Res. 2019;79(13):3185–3191. doi: 10.1158/0008-5472.CAN-19-0542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ma H., Qi G., Han F., et al. HMGB3 promotes PARP inhibitor resistance through interacting with PARP1 in ovarian cancer. Cell Death Dis. 2022;13(3):263. doi: 10.1038/s41419-022-04670-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yu H., Kortylewski M., Pardoll D. Crosstalk between cancer and immune cells: role of STAT3 in the tumour microenvironment. Nat Rev Immunol. 2007;7(1):41–51. doi: 10.1038/nri1995. [DOI] [PubMed] [Google Scholar]
- 91.Zou S., Tong Q., Liu B., et al. Targeting STAT3 in cancer immunotherapy. Mol Cancer. 2020;19:145. doi: 10.1186/s12943-020-01258-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ding L., Chen X., Xu X., et al. PARP1 suppresses the transcription of PD-L1 by poly(ADP-ribosyl)ating STAT3. Cancer Immunol Res. 2019;7(1):136–149. doi: 10.1158/2326-6066.CIR-18-0071. [DOI] [PubMed] [Google Scholar]
- 93.Martincuks A., Song J., Kohut A., et al. PARP inhibition activates STAT3 in both tumor and immune cells underlying therapy resistance and immunosuppression in ovarian cancer. Front Oncol. 2021;11 doi: 10.3389/fonc.2021.724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Färkkilä A., Rodríguez A., Oikkonen J., et al. Heterogeneity and clonal evolution of acquired PARP inhibitor resistance in TP53- and BRCA1-deficient cells. Cancer Res. 2021;81(10):2774–2787. doi: 10.1158/0008-5472.CAN-20-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hammond E.M., Asselin M.C., Forster D., et al. The meaning, measurement and modification of hypoxia in the laboratory and the clinic. Clin Oncol. 2014;26(5):277–288. doi: 10.1016/j.clon.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 96.Brown J.M. Exploiting the hypoxic cancer cell: mechanisms and therapeutic strategies. Mol Med Today. 2000;6(4):157–162. doi: 10.1016/s1357-4310(00)01677-4. [DOI] [PubMed] [Google Scholar]
- 97.Mehibel M., Xu Y., Li C.G., et al. Eliminating hypoxic tumor cells improves response to PARP inhibitors in homologous recombination-deficient cancer models. J Clin Invest. 2021;131(11) doi: 10.1172/JCI146256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chan N., Pires I.M., Bencokova Z., et al. Contextual synthetic lethality of cancer cell kill based on the tumor microenvironment. Cancer Res. 2010;70(20):8045–8054. doi: 10.1158/0008-5472.CAN-10-2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gachechiladze M., Škarda J., Soltermann A., et al. RAD51 as a potential surrogate marker for DNA repair capacity in solid malignancies. Int J Cancer. 2017;141(7):1286–1294. doi: 10.1002/ijc.30764. [DOI] [PubMed] [Google Scholar]
- 100.Castroviejo-Bermejo M., Cruz C., Llop-Guevara A., et al. A RAD51 assay feasible in routine tumor samples calls PARP inhibitor response beyond BRCA mutation. EMBO Mol Med. 2018;10(12) doi: 10.15252/emmm.201809172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pellegrino B., Herencia-Ropero A., Llop-Guevara A., et al. Preclinical in vivo validation of the RAD51 test for identification of homologous recombination-deficient tumors and patient stratification. Cancer Res. 2022;82(8):1646–1657. doi: 10.1158/0008-5472.CAN-21-2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liu Y., Burness M.L., Martin-Trevino R., et al. RAD51 mediates resistance of cancer stem cells to PARP inhibition in triple-negative breast cancer. Clin Cancer Res. 2017;23(2):514–522. doi: 10.1158/1078-0432.CCR-15-1348. [DOI] [PubMed] [Google Scholar]
- 103.Heeke A.L., Pishvaian M.J., Lynce F., et al. Prevalence of homologous recombination-related gene mutations across multiple cancer types. JCO Precis Oncol. 2018 doi: 10.1200/PO.17.00286. 2018:PO.17.00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ipsen M.B., Sørensen E.M.G., Thomsen E.A., et al. A genome-wide CRISPR-Cas9 knockout screen identifies novel PARP inhibitor resistance genes in prostate cancer. Oncogene. 2022;41(37):4271–4281. doi: 10.1038/s41388-022-02427-2. [DOI] [PubMed] [Google Scholar]
- 105.Peng Y., Liao Q., Tan W., et al. The deubiquitylating enzyme USP15 regulates homologous recombination repair and cancer cell response to PARP inhibitors. Nat Commun. 2019;10:1224. doi: 10.1038/s41467-019-09232-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sun C., Fang Y., Yin J., et al. Rational combination therapy with PARP and MEK inhibitors capitalizes on therapeutic liabilities in RAS mutant cancers. Sci Transl Med. 2017;9(392) doi: 10.1126/scitranslmed.aal5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yang B., Li X., Fu Y., et al. MEK inhibition remodels the immune landscape of mutant KRAS tumors to overcome resistance to PARP and immune checkpoint inhibitors. Cancer Res. 2021;81(10):2714–2729. doi: 10.1158/0008-5472.CAN-20-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Paes Dias M., Tripathi V., van der Heijden I., et al. Loss of nuclear DNA ligase III reverts PARP inhibitor resistance in BRCA1/53BP1 double-deficient cells by exposing ssDNA gaps. Mol Cell. 2021;81(22):4692–4708. doi: 10.1016/j.molcel.2021.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Taglialatela A., Leuzzi G., Sannino V., et al. REV1-Polζ maintains the viability of homologous recombination-deficient cancer cells through mutagenic repair of PRIMPOL-dependent ssDNA gaps. Mol Cell. 2021;81(19):4008–4025. doi: 10.1016/j.molcel.2021.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chen S.H., Yu X. Targeting dePARylation selectively suppresses DNA repair-defective and PARP inhibitor-resistant malignancies. Sci Adv. 2019;5(4) doi: 10.1126/sciadv.aav4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Parmar K., Kochupurakkal B.S., Lazaro J.B., et al. The CHK1 inhibitor prexasertib exhibits monotherapy activity in high-grade serous ovarian cancer models and sensitizes to PARP inhibition. Clin Cancer Res. 2019;25(20):6127–6140. doi: 10.1158/1078-0432.CCR-19-0448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhou S., Li D., Xiao D., et al. Inhibition of PKM2 enhances sensitivity of olaparib to ovarian cancer cells and induces DNA damage. Int J Biol Sci. 2022;18(4):1555–1568. doi: 10.7150/ijbs.62947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kanakkanthara A., Hou X., Ekstrom T.L., et al. Repurposing ceritinib induces DNA damage and enhances PARP inhibitor responses in high-grade serous ovarian carcinoma. Cancer Res. 2022;82(2):307–319. doi: 10.1158/0008-5472.CAN-21-0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gabbasov R., Benrubi I.D., O'Brien S.W., et al. Targeted blockade of HSP90 impairs DNA-damage response proteins and increases the sensitivity of ovarian carcinoma cells to PARP inhibition. Cancer Biol Ther. 2019;20(7):1035–1045. doi: 10.1080/15384047.2019.1595279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang Q., Bergholz J.S., Ding L., et al. STING agonism reprograms tumor-associated macrophages and overcomes resistance to PARP inhibition in BRCA1-deficient models of breast cancer. Nat Commun. 2022;13:3022. doi: 10.1038/s41467-022-30568-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Li L., Karanika S., Yang G., et al. Androgen receptor inhibitor-induced “BRCAness” and PARP inhibition are synthetically lethal for castration-resistant prostate cancer. Sci Signal. 2017;10(480) doi: 10.1126/scisignal.aam7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fugger K., Bajrami I., Silva Dos Santos M., et al. Targeting the nucleotide salvage factor DNPH1 sensitizes BRCA-deficient cells to PARP inhibitors. Science. 2021;372(6538):156–165. doi: 10.1126/science.abb4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Li S., Wang L., Wang Y., et al. The synthetic lethality of targeting cell cycle checkpoints and PARPs in cancer treatment. J Hematol Oncol. 2022;15:147. doi: 10.1186/s13045-022-01360-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gupta N., Huang T.T., Horibata S., et al. Cell cycle checkpoints and beyond: exploiting the ATR/CHK1/WEE1 pathway for the treatment of PARP inhibitor–resistant cancer. Pharmacol Res. 2022;178 doi: 10.1016/j.phrs.2022.106162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kim H., Xu H., George E., et al. Combining PARP with ATR inhibition overcomes PARP inhibitor and platinum resistance in ovarian cancer models. Nat Commun. 2020;11:3726. doi: 10.1038/s41467-020-17127-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sharma R.A., Plummer R., Stock J.K., et al. Clinical development of new drug-radiotherapy combinations. Nat Rev Clin Oncol. 2016;13(10):627–642. doi: 10.1038/nrclinonc.2016.79. [DOI] [PubMed] [Google Scholar]
- 122.Veuger S.J., Hunter J.E., Durkacz B.W. Ionizing radiation-induced NF-kappaB activation requires PARP-1 function to confer radioresistance. Oncogene. 2009;28(6):832–842. doi: 10.1038/onc.2008.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Boudra M.T., Bolin C., Chiker S., et al. PARP-2 depletion results in lower radiation cell survival but cell line-specific differences in poly(ADP-ribose) levels. Cell Mol Life Sci. 2015;72(8):1585–1597. doi: 10.1007/s00018-014-1765-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mao Y., Huang X., Shuang Z., et al. PARP inhibitor olaparib sensitizes cholangiocarcinoma cells to radiation. Cancer Med. 2018;7(4):1285–1296. doi: 10.1002/cam4.1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bi Y., Verginadis I.I., Dey S., et al. Radiosensitization by the PARP inhibitor olaparib in BRCA1-proficient and deficient high-grade serous ovarian carcinomas. Gynecol Oncol. 2018;150(3):534–544. doi: 10.1016/j.ygyno.2018.07.002. [DOI] [PubMed] [Google Scholar]
- 126.Mangoni M., Sottili M., Salvatore G., et al. Enhancement of soft tissue sarcoma cell radiosensitivity by poly(ADP-ribose) polymerase-1 inhibitors. Radiat Res. 2018;190(5):464–472. doi: 10.1667/RR15035.1. [DOI] [PubMed] [Google Scholar]
- 127.Abbotts R., Topper M.J., Biondi C., et al. DNA methyltransferase inhibitors induce a BRCAness phenotype that sensitizes NSCLC to PARP inhibitor and ionizing radiation. Proc Natl Acad Sci U S A. 2019;116(45):22609–22618. doi: 10.1073/pnas.1903765116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wilson A., Menon V., Khan Z., et al. Nitric oxide-donor/PARP-inhibitor combination: a new approach for sensitization to ionizing radiation. Redox Biol. 2019;24 doi: 10.1016/j.redox.2019.101169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Verhagen C.V.M., de Haan R., Hageman F., et al. Extent of radiosensitization by the PARP inhibitor olaparib depends on its dose, the radiation dose and the integrity of the homologous recombination pathway of tumor cells. Radiother Oncol. 2015;116(3):358–365. doi: 10.1016/j.radonc.2015.03.028. [DOI] [PubMed] [Google Scholar]
- 130.Sizemore S.T., Mohammad R., Sizemore G.M., et al. Synthetic lethality of PARP inhibition and ionizing radiation is p53-dependent. Mol Cancer Res. 2018;16(7):1092–1102. doi: 10.1158/1541-7786.MCR-18-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Riley R.S., June C.H., Langer R., et al. Delivery technologies for cancer immunotherapy. Nat Rev Drug Discov. 2019;18(3):175–196. doi: 10.1038/s41573-018-0006-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wang Z., Sun K., Xiao Y., et al. Niraparib activates interferon signaling and potentiates anti-PD-1 antibody efficacy in tumor models. Sci Rep. 2019;9:1853. doi: 10.1038/s41598-019-38534-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lassen U. Combining PARP inhibition with PD-1 inhibitors. Lancet Oncol. 2019;20(9):1196–1198. doi: 10.1016/S1470-2045(19)30509-1. [DOI] [PubMed] [Google Scholar]
- 134.Vinayak S., Tolaney S.M., Schwartzberg L., et al. Open-label clinical trial of niraparib combined with pembrolizumab for treatment of advanced or metastatic triple-negative breast cancer. JAMA Oncol. 2019;5(8):1132–1140. doi: 10.1001/jamaoncol.2019.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Konstantinopoulos P.A., Waggoner S., Vidal G.A., et al. Single-arm phases 1 and 2 trial of niraparib in combination with pembrolizumab in patients with recurrent platinum-resistant ovarian carcinoma. JAMA Oncol. 2019;5(8):1141–1149. doi: 10.1001/jamaoncol.2019.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sandhu S.K., Hussain M., Mateo J., et al. PROfound: phase III study of olaparib versus enzalutamide or abiraterone for metastatic castration-resistant prostate cancer (mCRPC) with homologous recombination repair (HRR) gene alterations. Ann Oncol. 2019;30:ix188–ix189. [Google Scholar]
- 137.Yap T.A., Bessudo A., Hamilton E., et al. IOLite: phase 1b trial of doublet/triplet combinations of dostarlimab with niraparib, carboplatin-paclitaxel, with or without bevacizumab in patients with advanced cancer. J Immunother Cancer. 2022;10(3) doi: 10.1136/jitc-2021-003924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Powles T., Carroll D., Chowdhury S., et al. An adaptive, biomarker-directed platform study of durvalumab in combination with targeted therapies in advanced urothelial cancer. Nat Med. 2021;27(5):793–801. doi: 10.1038/s41591-021-01317-6. [DOI] [PubMed] [Google Scholar]
- 139.Färkkilä A., Gulhan D.C., Casado J., et al. Immunogenomic profiling determines responses to combined PARP and PD-1 inhibition in ovarian cancer. Nat Commun. 2020;11(1):1459. doi: 10.1038/s41467-020-15315-8. [DOI] [PMC free article] [PubMed] [Google Scholar]