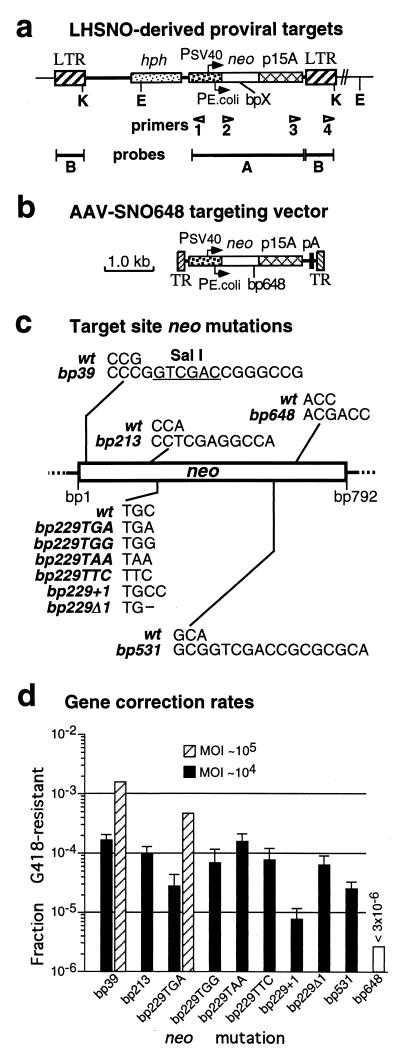
FIG. 1.
Gene correction of retroviral vector target loci. (a) Map of integrated proviral target sites based on LHSNO retroviral vector. The positions of sequencing primers, Southern analysis probes, and KpnI (K) and EcoRI (E) restriction sites are shown. bpX indicates the presence of the neo mutations shown in panel c. (b) Map of AAV-SNO648 targeting vector (12) containing the bp648 neo mutation. The locations of AAV terminal repeats (TR), SV40 (Psv40) and Tn5 (Pe.coli) promoters, transcriptional start sites (arrows), hph and neo genes, p15A plasmid replication origin, polyadenylation site (pA), and retrovirus LTRs (LTR) are shown in panels a and b. (c) Sequences of the wild-type (wt) neo gene present in retroviral vector LHSNO and the different neo mutations present in LHSNO-derived proviral target sites. Mutation names are based on the nucleotide position within the 792-bp neo coding sequence. A SalI restriction site is indicated (underlined bases). (d) Correction rates of the indicated neo mutations in HT-1080 cells containing LHSNO-derived target loci, infected with the AAV-SNO648 vector at an MOI of 104 (solid columns; bars indicate standard errors; n = 3) or 105 (hatched columns; n = 1) vector particles/cell, then plated in the presence or absence of G418. The fraction G418-resistant is the number of G418-resistant colonies/the total number of (unselected) colonies (see Materials and Methods). No G418-resistant colonies were obtained with cells containing the bp648 neo mutation at an MOI of 104 (open column). The reversion rates of all the polyclonal populations were <2 × 10−7, except for those containing the bp229TGG mutation, which had a reversion rate of 2.6 × 10−6 (still below the correction rate).