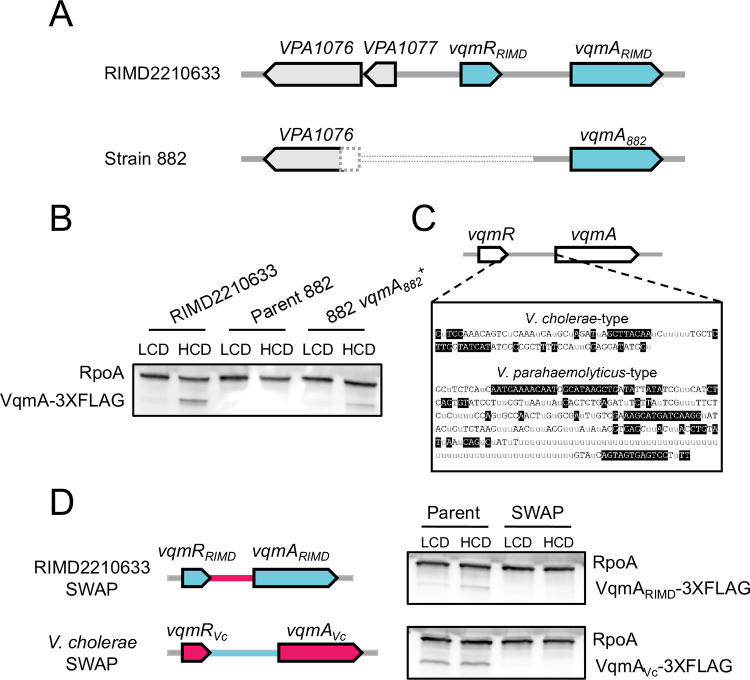
Fig 2. VqmA and VqmR are not produced in strain 882.
(A) Schematic of the vqmR-vqmA loci of V. parahaemolyticus RIMD2210633 and strain 882. The dotted pattern signifies that strain 882 harbors an 897 bp deletion encompassing a portion of a neighboring upstream gene (VPA1076, encoding a putative transcriptional regulator of a proline metabolic operon), a putative gene of unknown function (VPA1077), the vqmR promoter, the vqmR coding sequence, and a portion of the non-coding region upstream of vqmA882. (B) Representative western blot showing VqmARIMD-3XFLAG in V. parahaemolyticus RIMD2210633, VqmA882-3XFLAG in strain 882, and VqmA882-3XFLAG in strain 882 with the non-coding region upstream of vqmA882 restored (i.e., this strain is 882 vqmA882+). RpoA was used as the loading control. (C) Consensus sequences of the intergenic regions between vqmR and vqmA for the two designated groups. Gray “u” letters indicate ≤50% agreement. Black nucleotides with clear backgrounds indicate >50% but <100% agreement. White nucleotides with black backgrounds indicate 100% agreement. (D) Left: schematic showing exchange of regions between V. parahaemolyticus RIMD2210633 and V. cholerae at the vqmR-vqmA loci. Genes and non-coding regions from V. parahaemolyticus RIMD2210633 and V. cholerae are colored cyan and red, respectively. Right: representative western blot of VqmA-3XFLAG produced by V. parahaemolyticus RIMD2210633 and V. cholerae from their native promoters (Parent) and following exchange of their promoters (SWAP). RpoA was used as the loading control. Data are representative of three independent experiments (B) and two independent experiments (D).