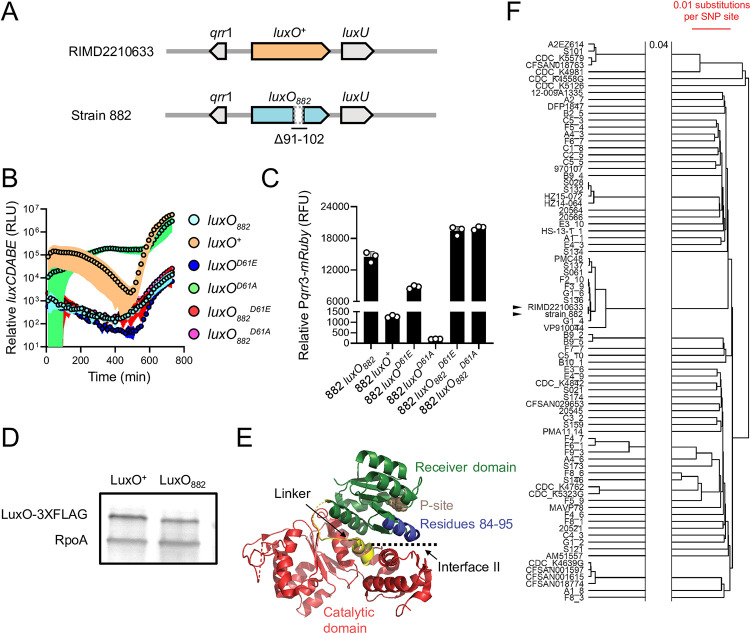
Fig 3. LuxO882 harbors a 12 amino acid deletion, rendering the protein LCD-locked.
(A) Schematic of the luxO-containing loci of V. parahaemolyticus RIMD2210633 and V. parahaemolyticus strain 882. Strain 882 luxO harbors a 36 bp deletion corresponding to residues 91–102, as indicated. (B) Relative luxCDABE output over time from the parent strain 882 (luxO882; cyan), strain 882 luxO+ (orange), strain 882 luxOD61E (blue), strain 882 luxOD61A (green), strain 882 luxO882D61E (red), and strain 882 luxO882D61A (purple). RLU designates relative light units. As noted in the main text, all luxO882 alleles harbor the 12 amino acid Δ91–102 deletion that is not present in the RIMD2210633 luxO alleles. (C) Relative Pqrr3-mRuby output for the indicated 882 strains. RFU designates relative fluorescence units. (D) Representative western blot of LuxO+-3XFLAG and LuxO882-3XFLAG produced by strain 882. RpoA was used as the loading control. (E) Crystal structure of V. angustum LuxO (PDB: 5EP0 and [16]). The receiver, catalytic, and linker domains are colored in green, red, and yellow, respectively. The aspartate site of phosphorylation is shown in brown (denoted P-site). Residues 84–95 in V. angustum LuxO, corresponding to residues 91–102 in V. parahaemolyticus LuxO, which are missing in V. parahaemolyticus strain 882 LuxO, are colored in blue. Interface II is indicated. (F) A phylogenetic tree constructed using UPGMA based on core genome SNP alignment of the 88 V. parahaemolyticus strains carrying the Δ91–102 LuxO mutation. Scale bar (red) indicates 0.01 nucleotide substitution per SNP site. The two horizontal lines indicate a gap in the tree, and 0.04 refers to nucleotide substitutions per SNP site separating the gap. Black arrowheads show strain 882 and V. parahaemolyticus RIMD2210633. Data are represented as means ± std with n = 3 biological replicates (B, C) and representative of three independent experiments (D).