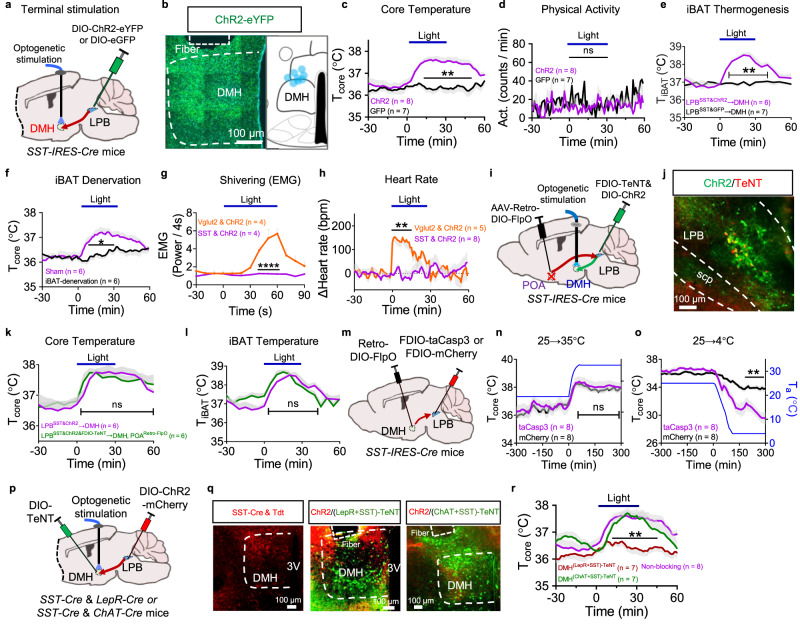
Fig. 7. The LPBSST → DMH projection is selectively required for iBAT thermogenesis during cold defense.
a, b Design to activate the LPBSST → DMH projection (a) and the representative expression of ChR2-eYFP (left) and summary of fiber tracts (shown as blue dots, right) in the DMH (b). This experiment was repeated at least 8 times independently with similar results. c–e Changes in Tcore (c), physical activity (d), and TiBAT (e) after photoactivation of the LPBSST → DMH projection. Animal numbers were indicated. f Denervation of the iBAT abolished the hyperthermia induced by photoactivation (n = 6 mice each). g, h Photoactivation of the LPBSST → DMH projection did not change nuchal muscle EMG (g; n = 4 mice each) and heart rate (h; SST, n = 8 mice; Vglut2, n = 5 mice). i Activating the LPBSST → DMH pathway while blocking POA-projecting LPBSST neurons using TeNT. j Representative expression of ChR2 (green) and TeNT (red) in the LPB. This experiment was repeated at least 6 times independently with similar results. k, l Changes in Tcore (k) and TiBAT (l) after photoactivation of LPBSST terminals in the DMH while blocking POA-projecting LPBSST neurons (n = 6 mice each). m Deleting DMH-projecting LPBSST neurons by neural killing with taCasp3. n, o Tcore changes during warm (n) and cold (o) exposures after deleting DMH-projecting LPBSST neurons (n = 8 mice each). p Photoactivation of LPBSST & ChR2 terminals in the DMH after blocking DMHLepR+SST neurons or DMHChAT+SST neurons. q Representative SST-Cre & Tdt (left panel) and LPBSST & ChR2 terminals (red) and TeNT (green) expression in the DMH (right two panels). This experiment was repeated at least 3 times independently with similar results. r Changes in Tcore after photoactivation of LPBSST terminal in the DMH while blocking DMHLepR+SST or DMHChAT+SST neurons (n = 7 mice each). All data are the mean ± sem, and (c–h, k, l, n, o, r) were analyzed by two-way RM ANOVA followed by Bonferroni’s multiple comparisons test. The p-values are calculated based on statistical tests in Supplementary Table 2. *p ≤ 0.05; **p ≤ 0.01; ****p ≤ 0.0001; ns, not significant. Source data are provided as a Source data file.