Extended Data Figure 6. Strand-seq validation of the full-coverage Verkko trio assembly and HPRC manually curated assembly 11.
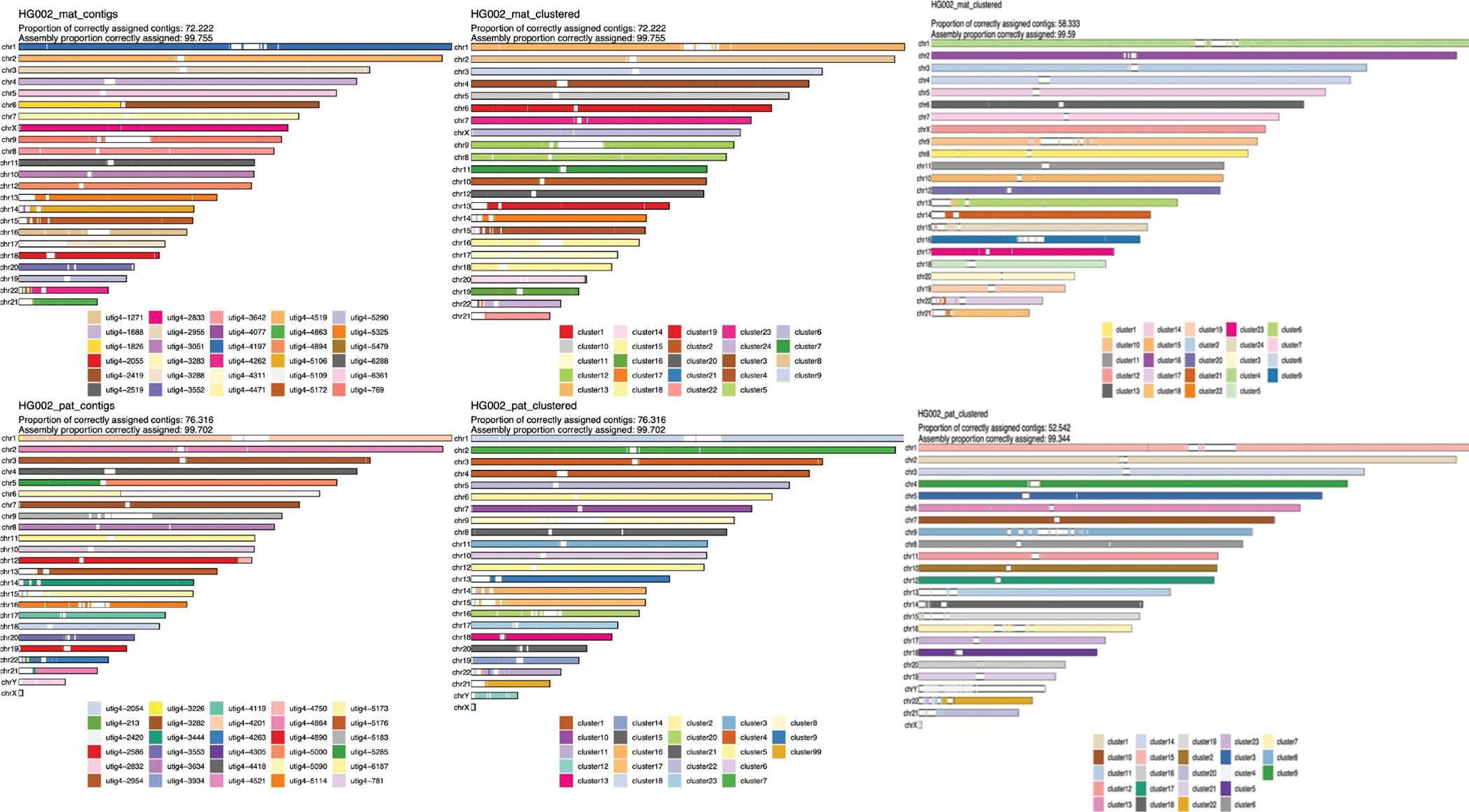
The maternal haplotype is shown along the top row and the paternal along the bottom row. Leftmost: alignment-based scaffold assignment to the maternal haplotype (top) and paternal haplotype (bottom) for the full-coverage Verkko assembly. Almost all chromosomes are a single color, indicating that Verkko scaffolds resolved most chromosomes end-to-end. The only exceptions are in the acrocentrics, where some of the scaffolds could not be assigned due to low mappability and maternal Chromosome 6 and paternal Chromosomes 5 which are each composed of two large scaffolds. Over 99.7% of the scaffold bases could be assigned to chromosomes. Middle: the cluster assignment for the maternal haplotype (top) and paternal haplotype (bottom) based on Strand-seq data for the full-coverage Verkko assembly. Here, cluster ID is assigned to each 200 kb window in a scaffold. In case of large scale chromosomal mis-joins, we expect to see multiple colors in a chromosome. The Verkko assembly is consistent with scaffolds all representing a single chromosome bin. Once again, >99.7% of the scaffold bases can be assigned using Strand-seq. Only 2 and 4 Mb of sequence not scaffolded by Verkko could be assigned to the maternal and paternal haplotypes, respectively. Right: The cluster assignment for the maternal haplotype (top) and the paternal haplotype (bottom) based on Strand-seq data for the HPRC manually curated assembly. Here, cluster ID is assigned to each 200 kb window in a scaffold. In case of large scale chromosomal mis-joins, we expect to see multiple colors in a chromosome. A smaller fraction of contigs (and a slightly lower fraction of bases) was assigned than for the Verkko assembly, despite the combination of technologies and manual curation. This may be due to shorter contigs from unresolved repeats which are resolved through Verkko’s ONT integration. There is also visible chromosome mixing within the acrocentric chromosomes unlike in the Verkko result.
