Extended Data Figure 7. Strand-seq structural variant analysis for Verkko full-coverage assembly.
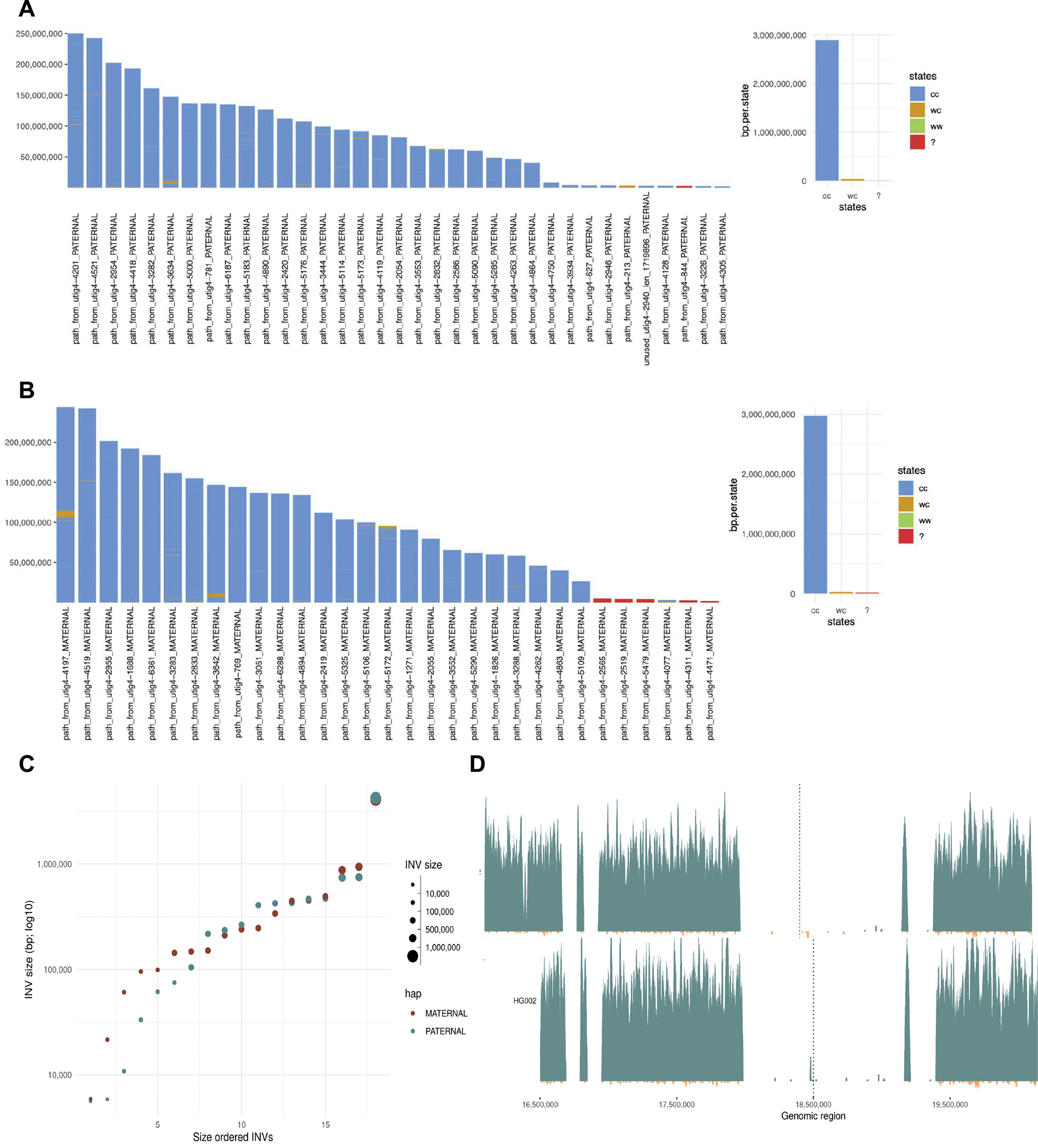
The states assigned to each scaffold in the paternal (A) and maternal (B) for the full-coverage Verkko trio assembly. Strand-seq reads aligned to each assembly are genotype based on their directionality into three possible strand states. Crick-Crick (‘cc’) state in which both homologs in Strand-seq data map in direct orientation and thus such regions are consistent with Strand-seq directional information. Watson-Watson (‘ww’) state in which both homologs in Strand-seq data map in inverted orientation and are indicative of assembly misorientation or unresolved homozygous inversion. Lastly, there are a few (<1% of bases) Watson-Crick (‘wc’) where there is a mixture of Watson and Crick reads and such regions are indicative of heterozygous inversions between haplotypes or might also be cause by low-mappability regions for short Strand-seq reads. C. The size of the heterozygous inversion versus the count of inversions of that size in the maternal and paternal haplotypes of the full-coverage Verkko trio assembly. These regions have confident Strand-seq alignments and normal copy number so these regions indicate potential true heterozygous variation between the haplotypes. D. Strand-seq alignments to the reference Chromosome Y before it was corrected (top) and full-coverage Verkko trio Chromosome Y assembly (bottom). Each plot shows Strand-seq directional read coverage reported as binned (bin size: 10,000, step size: 1,000) read counts represented as vertical bars above (teal; Crick read counts) and below (orange; Watson read counts) the midline. The top plot shows an inversion (dashed line) where directly oriented reads (Crick; teal) switch to inversely oriented reads (Watson, orange) and then back to directly oriented reads. The Verkko assembly in contrast is consistent with only Crick reads present in the same location (dashed line).
