Figure 3. Verkko assembly graph of the HG002 diploid genome.
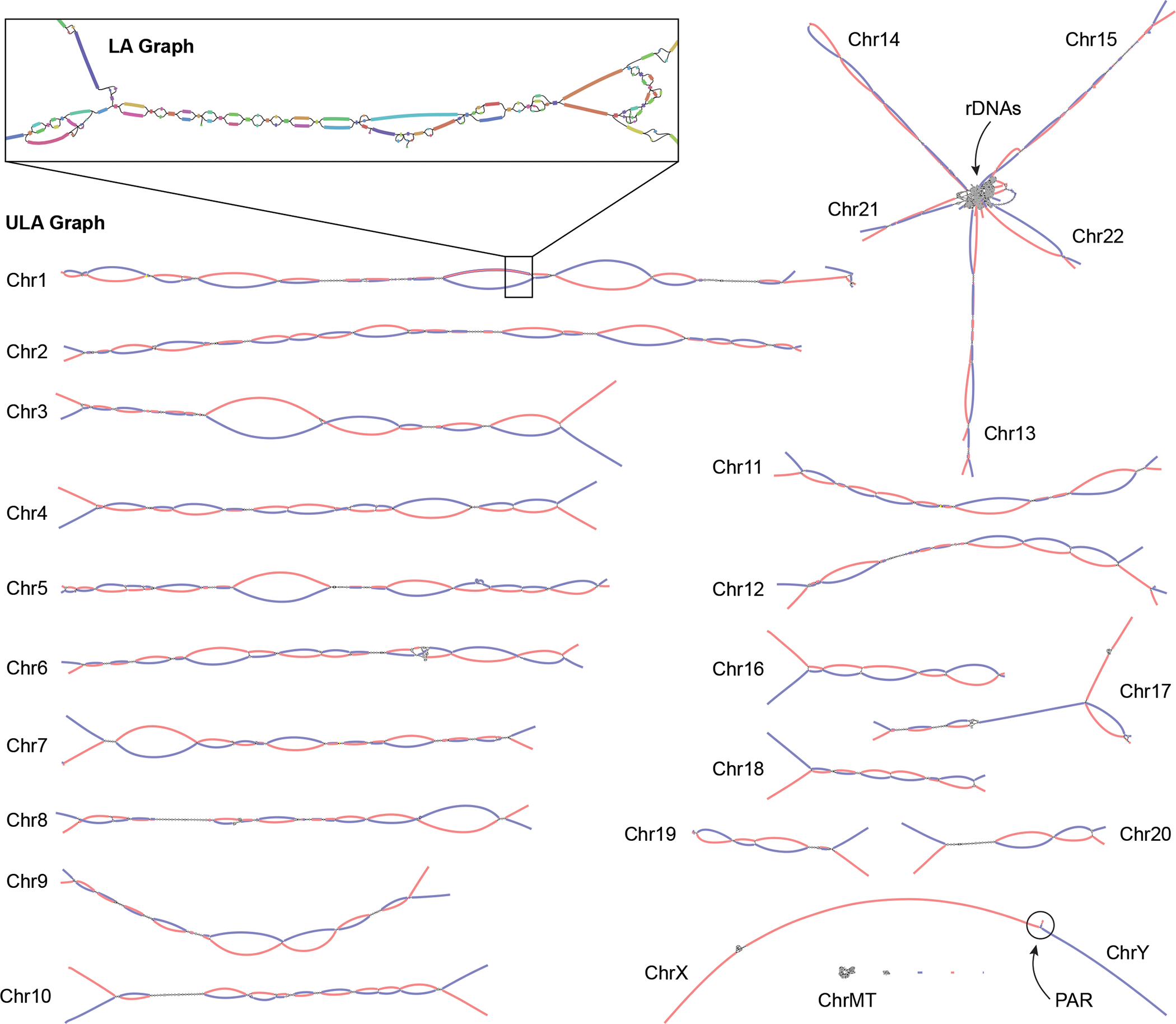
ULA graph integrating both PacBio HiFi and ONT ultra-long reads visualized using Bandage62. Inset shows a portion of the LA graph with multiple bubbles, tangles, and tips prior to resolution using the ultra-long reads. This particular region contains stretches of homozygosity and exact repeats that could not be resolved by HiFi alone. After integration of the ONT data, the corresponding region in the ULA graph is resolved into two linear haplotypes. Each chromosome in HG002 is mostly resolved as a single connected component, with the exception of the acrocentric (13, 14, 15, 21, and 22) and sex chromosomes (X and Y), which are joined by the highly similar rDNA arrays and pseudoautosomal regions (PAR), respectively. The final unitigs in the ULA graph have been colored after assembly by trio-derived haplotype markers (maternal, red; paternal, blue). Using this information to determine haplotype paths through the graph, Verkko was able to completely assemble 20 chromosomal haplotypes in HG002 from telomere to telomere without gaps.
