Extended Data Figure 2. Verkko CHM13 assembly sub-graphs.
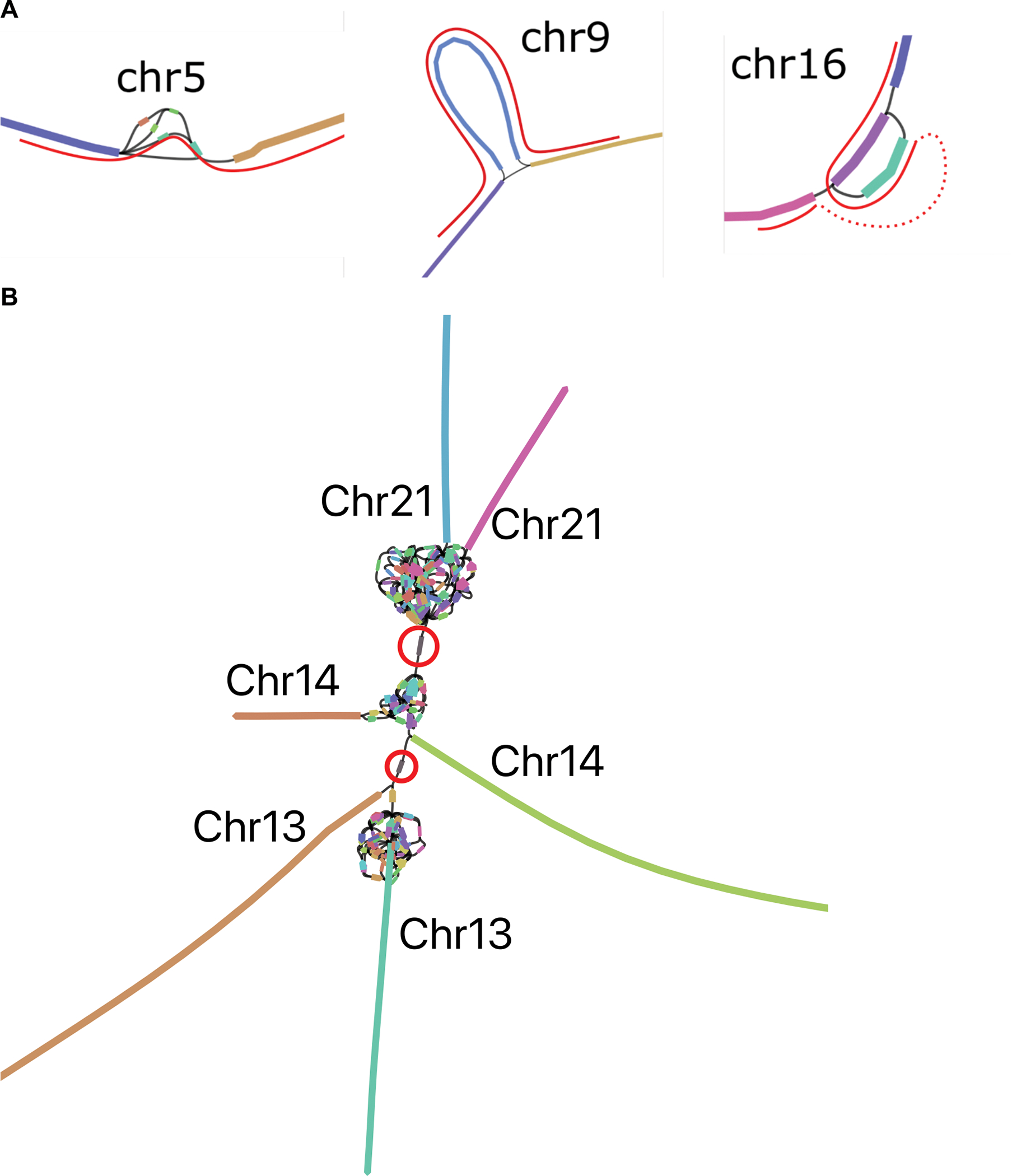
A. The remaining unresolved regions in CHM13 chromosomes 5, 9 and 16, visualized using Bandage62, with the correct resolution marked in red paths. Left: Chr5 has a spurious edge causing a cycle, and three spurious low-coverage nodes which were not removed by bubble popping since they are a part of the cycle. Middle: Chr9 has a spurious edge. Right: Chr16 has two spurious edges, and one missing edge (dashed red curve). The spurious non-genomic edges are caused by noisy ONT alignments switching between highly similar repeats in the LA graph, while the missing edge is caused by low HiFi coverage. B. rDNA cluster mixing in CHM13 chromosomes 13, 14, and 21, visualized using Bandage62. Each chromosome has a separate rDNA tangle. There are two cross-chromosomal connections by erroneous low coverage (<4x) nodes circled in red. For all three chromosomes, the remainder of the p and q arms are contained in the long unitigs shown.
