Extended Data Figure 5. IGV 82 views of a recently published HG002 diploid assembly of paternal Chromosome 10 11 (top) and the Verkko full-coverage trio assembly of the same chromosome (bottom).
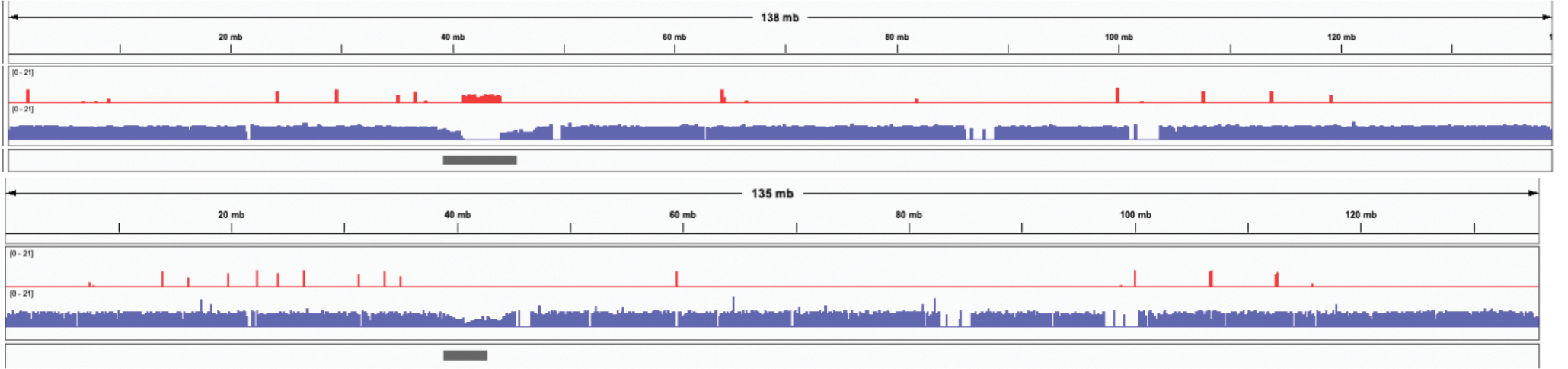
The tracks show the maternal (red) and paternal (blue) markers. The centromere location is shown in gray. The published assembly has extensive switching within the centromere array, indicated by the presence of maternal markers and the absence of paternal markers. In contrast, the Verkko assembly centromere shows only paternal markers. The Verkko paternal centromere array is shorter but shows no signs of mis-assembly (Extended Data Figure 8) indicating the larger array in the published assembly is likely due to the incorrect insertion of maternal sequence. Overall, the Verkko assembly is more continuous, with 0 gaps vs 4, and a lower hamming error rate, 0.03%, versus 1.98% compared to the published assembly.
